Research lines Molecular Cytology
-
High resolution microscopy - Prof. Dorus Gadella
It is well known that there is a spatial limit to which light can focus: approximately half of the wavelength of the light you are using. But this is not a true barrier, because this diffraction limit is only true in the far-field and localisation precision can be increased with many photons and careful analysis. The image of a point source on a microscope detector is called the point-spread function (PSF), which is limited by diffraction to be approximately half the wavelength of the light. But it is possible to simply fit that PSF with a Gaussian to locate the center of the PSF, and thus the location of the fluorophore with a much higher accuracy
Betzig et al. (Science) developed photo-activated localisation microscopy (PALM) while Zhuang and co-workers used a similar technique called stochastic optical reconstruction microscopy (STORM). In both techniques samples filled with many dark fluorophores are imaged. The dyes can be photoactivated into a fluorescing state by a flash of light. Because photoactivation is stochastic, only a few, well separated molecules "turn on". Then Gaussians are fit to their PSFs in order to localise the centre of the particle. After the few bright molecules photobleach (sometimes actively by using another differently colored excitation source), the next flash of the photoactivating light activates random fluorophores again and the PSFs are fit of these different molecules. This process is repeated many times, building up an image. Because the molecules were switched on-and-off (and thus localised) at different times, the 'resolution' of the final image can be much higher than that limited by diffraction. We are currently setting up a PALM microscope and applying this technique to study signal transduction pathways
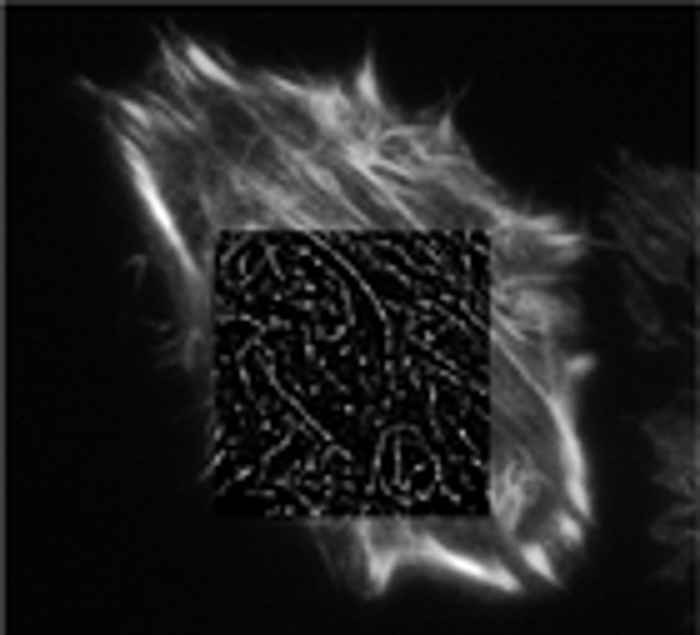
-
Spatial organisation of sub-cellular signalling - Dr Joachim Goedhart and Dr Mark Hink
By employing genetic encoded fluorescent biosensors we analyse the in situ molecular interactions between signalling molecules (phospholipid-second messengers, receptors, G-proteins and effector molecules) and flow of information across and in the plane of the membrane of living mammalian cells. We aim to understand how cells can achieve and maintain a local signal in the membrane (e.g. in order to drive morphogenesis, or to define new cytoskeletal anchorage or vesicle-docking sites).
-
Fluorescence fluctuation spectroscopy (FFS) to perform quantitative microscopy - Dr Mark Hink
One of the most intriguing challenges in life sciences is to understand how a complex mixture of molecular particles and structures can make up a living cell. Despite the immense number of studies still much is unknown about the molecular basis of numerous biological processes such as cell proliferation, differentiation, intra- and extra-cellular communication and apoptosis. To increase our understanding about the complexity of these processes in living cells, experimental and especially quantitative data on the spatial-temporal organisation is required. Fluorescence based techniques are ideal tools for this type of studies.
Fluorescence Fluctuation Spectroscopy (FFS) is a family of fluorescence techniques that is capable of detecting concentration, dynamics and interactions of fluorescent particles down to the single-molecule level and, if desired, in the living cell. We are applying, optimising and expanding FFS techniques like FCCS, PCH, PIE-FLCS, stICS and RICS to describe signal transduction pathways, like the Galpha signalling pathway quantitatively. Thereto, the proteins of interest are genetically labeled with the various color- and lifetime variants of the green fluorescent protein (partly developed in our laboratory) expressed in living cells and studied by advanced fluorescence microscopes.