Research Priority Areas (RPA's)
-
Green Life Sciences
author: prof. Michel Haring
Green Life Sciences aims to understand the origins and regulation of diversity at three levels (genomes, molecular signals and responses) underlying the flexibility of plants and their major pests and pathogens to changing environments. The development of sustainable strategies for crop and biodiversity management requires an integrated and multidisciplinary approach, combining studies on pest and pathogen resistance, plant adaptation, natural selection, ecological genomics, stress physiology and plant development.
Plants are fundamental to life on our planet. It is generally acknowledged that global changes in temperature, rainfall, soil fertility and air quality are becoming a major problem for agriculture and the provision of natural resources. It is a central scientific goal to understand the impact of these changes and how plants react to environmental change. This requires studying natural variation among and within species, so that we can determine which flexibility plants express when coping with these challenges. This also includes effects that result from the interaction with pathogens and pests, as the distribution and evolution of a plant’s enemies is also affected by environmental change.

- Feasible to predict
If we know which genes and molecules are essential to cope with new environments it becomes feasible to predict how plants will respond, paving the way for sustainable solutions to conserve earth’s biodiversity, as well as for innovations towards sustainable food production.
- Joint expertise
The joint expertise within the Swammerdam Institute of Life Sciences and the Institute of Biodiversity and Ecosystem Dynamics in combination with external partners (VU, plant breeding industry) forms an ideal cluster to tackle these issues.

-
Systems Biology
author: prof. Age Smilde
Research Priority Area Systems Biology: Modeling host-microbiome interactions.
How do microbes interact with their host? What kind of direct and indirect interactions are present and can these be measured and modeled? In the Research Priority Area Systems Biology (RPA-SB) a group of researchers is answering these questions. The RPA-SB is a faculty sponsored research activity involving groups of SILS and IBED with shared PhDs and assistant professors. The biological systems studied are diverse. Within SILS these are mainly the rhizosphere/plant-host and the gut microbiome/human-host system. In IBED the studied system is mainly algae and their microbes. However, the type of questions and modeling are evolving around networks of interactions of microbes with their hosts and are all thus very similar. Due to the availability of advanced instrumentation we are now facing biological questions which go far beyond analyzing and modeling simple data sets. The figure shows examples of the interactions that take place in the plant-rhizosphere microbiome system. Modeling such a system clearly asks for a multidisciplinary approach. First, a proper experimental design has to be constructed carefully focusing on concrete parts of the system that need to be studied. Next, experiments have to be performed involving different types of technologies such as metabolomics, RNAseq gene-expression analysis and microbiome measurements such as Amplicon sequencing or shotgun metagenomics. After curation of the data and initial exploratory data analysis and bioinformatics, systems biology models need to be formulated and fitted to the data. This may result in biological findings which then need to be validated experimentally. Evidently, this is a long-term endeavor which cannot be concluded overnight!
-
Urban Mental Health
author: prof. Paul Lucassen
SILS is partner in the Center for Urban Mental Health (UMH), one of the main Research Priority Areas (RPA) of the University of Amsterdam. Its multidisciplinary research is embedded within the Faculties of Social and Behavioural Sciences (FMG), Medicine (A-UMC), Science (FNWI) and the Institute for Advanced Study (IAS).
Within UMH, state-of-the art complexity science is used as a backbone to understand and intervene upon the complexities and dynamics of mental health problems in an urban environment. Special attention is paid to common mental health disorders in the city, like anxiety, depression and addiction. UMH research is aimed at understanding why and when some groups or individuals thrive in an urban setting, whereas others are vulnerable and develop mental problems.
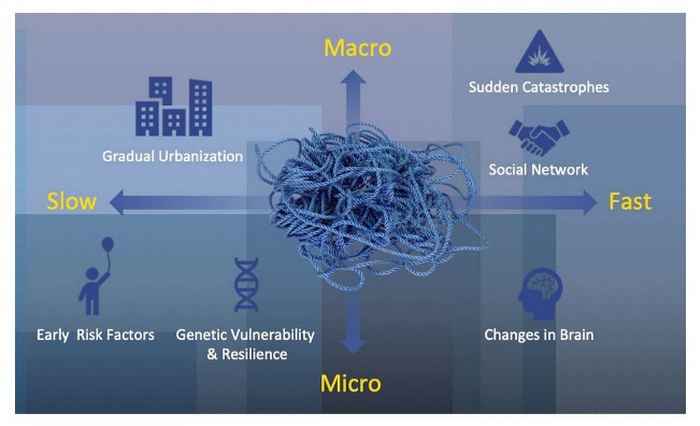
The Centre's approach integrates previously unconnected theories and sources, and thereby aims to unravel new pathways to improve urban mental health but now taking into account the many dimensions, complexities and dynamics of mental health in an urban environment. Ultimately, research within UMH aims to leverage new points for intervention as well as for policy making, that will be tested in collaboration with societal partners and will have direct relevance for mental health in general, and for Amsterdam in particular.
-
Personal Microbial Health
author: prof. Stanley Brul
This Research Priority Area was borne from the notion that our microbiome is a personal trait, shaped from birth, by an interplay of genetic, immunological, cultural, socio-economic, behavioral and environmental factors. As such the RPA formed the basis for the NWA-ORC METAHEALTH project that was later granted. We see microbiome health as a complex and personalized multiplex network of these processes actively sustaining health of the individual. Microbiome disbalance with the host (dysbiosis of the microbiota, the microbes, with their associated genomes) at a specific site, such as the oral cavity or gut, has implications for the entire body and can have severe consequences for general health. Therefore, delineating and understanding of these complex host-microbiome-environment interactions is of high importance for maintaining health and the prevention of disease.
Approach
The PMH RPA project was set-up from ACTA together with the Science Faculty and the Amsterdam UMCs’ medical faculty of the UvA. SILS provided its main expertise from the pre-existing Systems Biology of Host-Microbiome Interactions RPA. Work in PMH focuses on acquiring an interdisciplinary expertise in microbiomics, metagenomic analysis of all genomes in the microbiota of a specific body niche, among the RPA members. This means interdisciplinary seminars and courses covering biological, technical and socio-cultural aspects of microbiome science. This asks for an innovative approach to deal with the commonly occurring challenge of high heterogeneity in microbiome study protocols, outputs and data management. The aim in PMH is to unify and harmonize epidemiological, clinical, laboratory and bioinformatics approaches for studies involving microbiota (all the microorganisms in a given host niche) and their microbiomes. PMH enriches data from existing and ongoing cohorts (HELIUS https://www.heliusstudy.nl/nl/over-helius, AIMS https://aimsonderzoek.nl/, Sarphati ethnographic cohort https://www.sarphaticohort.nl/en/) with additional data on the systems biology of host-microbiome interactions and its socio-cultural context. The observations are integrated in microbiome personal health in-silico prediction models that we aim to apply for the generation of new hypotheses and for the prediction of general health outcomes. The data derived models are to be used for in silico simulation (Digital Twins) and modelling of microbiome resilience towards dysbiosis and discovery of preventive and therapeutic options for maintaining microbiome health.
-
Amsterdam Brain and Cognition
author: dr. Harm Krugers
Who are we? How do we take decisions? How do we process and store information, and how does this influence our behavior? What is consciousness? How do genes and environment interact and determine brain development and risk for disease? Which brain processes are involved in addiction, phobias or compulsive behavior? These are just a few examples of the types of questions that have been asked for centuries by scholars from various backgrounds. More than ever, we can provide answers to these questions by using new techniques, but in particular through collaborations with scientists from different disciplines. This interdisciplinary approach is the key to unraveling the mysteries of the mind, and its relation with the brain. The University of Amsterdam has a strong tradition in research on brain and cognitive processes and researchers from these different traditions, also from the faculty of Sciences at UvA and SILS-CNS, are brought together in the Amsterdam Brain and Cognition center (ABC).
ABC is a network organization, and has currently around 350 affiliated members, ranging from PhD students to full professors within the UvA. ABC holds the Research Priority Areas (RPA) Brain and Cognition and facilitates cooperation and stimulates novel and promising research projects in the area of brain and cognition. Although this RPA will stop, ABC aims to keep connecting scientists from different disciplines. Its main roles have been to fund bottom-up interdisciplinary and inter-faculty research projects through its ‘ABC Project grant’ and ‘ABC Talent grant’ programmes; fund excellent incoming researchers that contribute to the scientific field(s) through the ‘ABC VIP grant’ programme; organize and/or (co-)facilitate UvA-internal events (lectures, colloquia, networking days, symposia, etc.) on research topics connected to its themes, and train the future generation of brain and cognitive scientists via a very successful Master Brain and Cognitive Sciences programme, an annual ABC Summer School and facilitating the teaching availability of faculty researchers to the programme.
-
Data Science Centre
author: dr. Frans van der Kloet
Incorporate deep-learning tools in data analysis
The data science centre (DSC) was founded with the mission to enhance the university’s research by developing, sharing, and promoting data science methods and technologies across faculties. The most ubiquitous tools out there are based on deep learning (e.g., neural networks) and within the DSC they are common ground for most researchers, each in their respective fields. In SILS (initially) two researchers (Marc Galland and Frans van der Kloet) were nominated to be an affiliate of the DSC and set out to investigate how these deep-learning tools could be deployed to tackle the seemingly trivial, but very time-consuming problems (and error prone) of counting objects on images, like nematodes or trichomes. The deep-learning answer to this are so-called computer-vision solutions e.g., transfer learning: by using a pretrained model, only few samples are needed to retrain the model to identify the new classes. It only takes 20 minutes (on a low spec computer) to retrain a model originally pre-trained to classify cars and benches to identify the nematodes on a petri dish (see figure).

Original model to identify benches/cars. The application of computer vision solutions is very promising. To minimize the amount of manual pre-processing of the images that is still required, it is investigated how data augmentation by computer generated images can be applied. This could also address the issue of (natural) class imbalances (e.g., different numbers per class in the images).
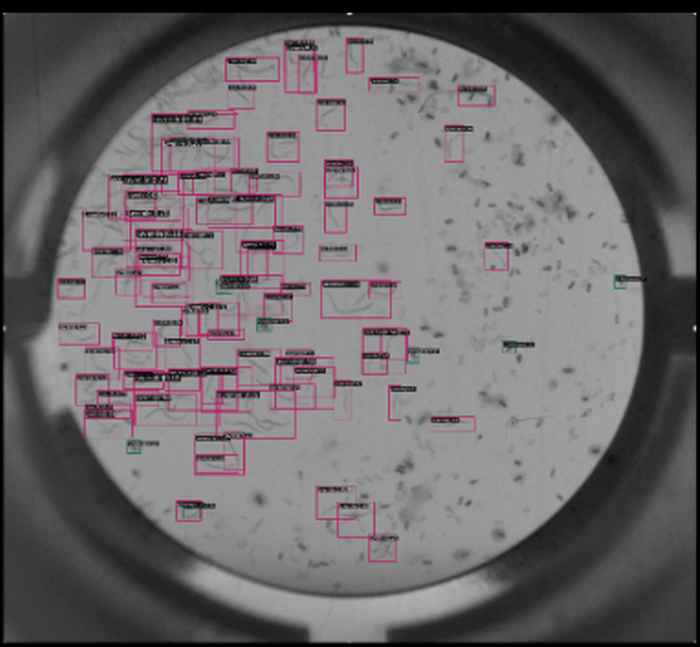
Retrained model to identify nematodes/eggs. With increasing complexity of the problem, the deep learning model also grows, limiting the applicability of applying these models to data analysis of biological samples where often only few samples exist. In addition, deep learning tools remain a black box most of times. Frans van der Kloet joined forces with Huub Hoefsloot and Andrew Foster (MSc student) to investigate the applicability of deep-learning tools in the analysis of biological systems. Inspired by visual networks (VNNs) (i.e., networks with pre-defined connections between nodes and layers) it was investigated how imposing sparseness penalties can be used to create meaningful and informative networks. The resulting method was applied to a dataset of RNA sequencing samples (~3,500) to predict 5 different cancer types. It successfully reduced a subset of 300 (pre-selected) genes to approximately 20 genes without loss of predictive power. Moreover, visualization of the knowledge graph (Figure 2) highlights genes of interest and possible hubs they group in. The work is still in progress and efforts will be made to scale the modelling up to start with 20k genes and validate how these hubs resemble known pathways. Currently, 3 people from SILS are affiliated with the DSC.