Using Epigenetics to Estimate Age: A New Tool for Forensic Investigations
UvA researchers combine X chromosome and autosomal epigenetic DNA methylation markers to refine age prediction of biological traces.
2 September 2025
"By integrating epigenetic sex chromosomal data with well-established autosomal markers, we can extract significantly more information, even when no profile match is available. This strengthens the link between molecular biology and forensic casework."
Even when a DNA profile can't be matched to a known person or found in a database, epigenetic age estimation can offer important clues to help the investigation move forward. Previous models focused mainly on autosomal markers. In this study, over 10,000 epigenetic DNA methylation sites from sex chromosomes were tested. The inclusion of a selected set of epigenetic X chromosomal markers combined with six autosomal ones, clearly improved prediction accuracy.
The research demonstrates how epigenetics, machine learning and bioinformatics can answer real-world forensic questions. The work builds on earlier research by Naue et al., also conducted within the same UvA research group. It illustrates how epigenetic DNA-based age estimation has evolved into a mature line of forensic inquiry, supported by machine learning and bioinformatics.
Published in Epigenetics & Chromatin (July 2025), the findings offer a valuable tool for age estimation from biological traces, particularly relevant in sexual and violent assault cases, unidentified human remains, and cold cases. This work was supported by the Co van Ledden Hulsebosch center (CLHC), the interdisciplinary expertise center for forensic scietific and medical research in the Netherlands.
Curious to work on these questions yourself? Explore the MSc Forensic Science and https://www.clhc.nl .
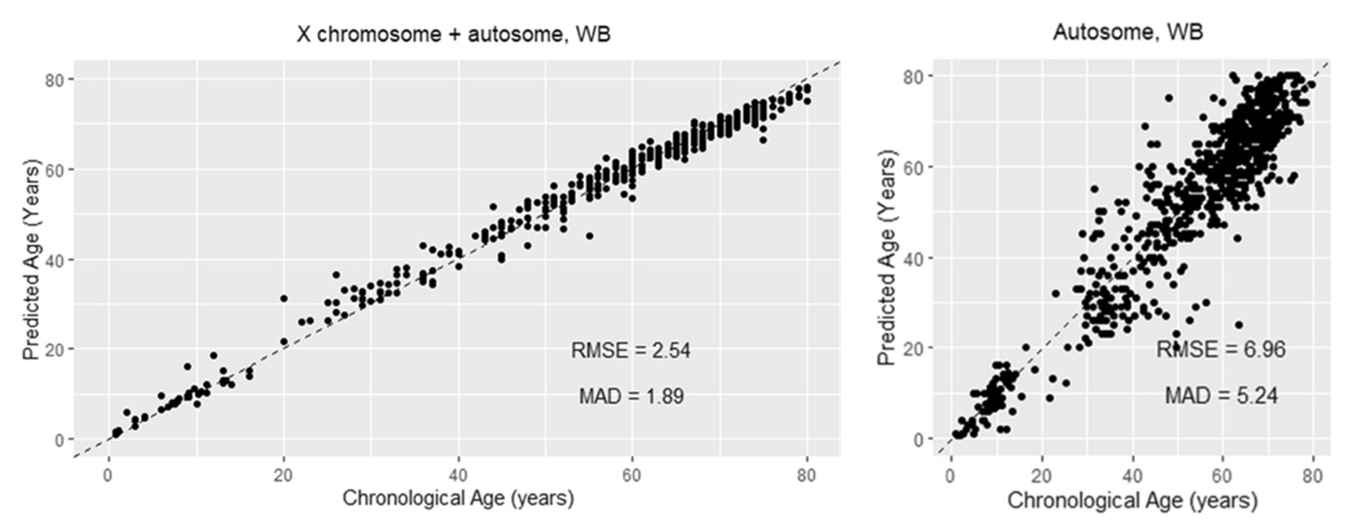
The figure shows a clear boost in accurracy to predict chronological age when including epigenetic blood markers from X chromosome and autosomal data.
The striking visual comparison in the left and right figure highlights the benefit of including X chromosomal epigenetic markers for DNA-based age prediction.
In the left figure, where predictions are based on a selected set of both X chromosome and autosomal DNA epigenetic markers, nearly all data points fall closely along the diagonal line: a sign that predicted ages closely match actual ages. In contrast, the right panel, which relies only on autosomal DNA epigenetic markers, shows a much wider scatter of points. In this case the predicted ages deviate significantly from the known age, and the error rate is clearly higher.
The visual takeaway: the integration of epigenetic X chromosomal data with epigenetic autosomal data leads to a noticeably more reliable epigenetic-based DNA age estimate.