Research lines RNA Biology & Applied Bioinformatics
-
Research line Dr R.J. (Rob) Dekker – smallRNA in disease and development
Because smallRNAs are the focus of much research worldwide, the decisive role of smallRNAs in disease and development is becoming increasingly evident.
smallRNA in disease
Since smallRNAs play an important role in disease processes, they also offer the chance to use them as biomarkers for diseases. An example of this is virus infections in plants.
Sometimes commercial plants harbor unwanted viruses. For many plant-breeding companies early and accurate detection of viruses in their breeding material is essential. Although several plant-virus detection methods, such as ELISA, are available, each of them has its own limitations. A major drawback is that one needs to know which virus is present to use the correct test and each virus requires a specific test. Another drawback is the fact that many detection methods identify the virus particles itself, whereas many viruses can “hide” themselves quite effectively. An easy method to detect the presence of any known virus anywhere in a plant would be very handy.
Silencing-RNAs (siRNAs) are small RNA molecules of 21-24 nucleotides that are abundantly present in plant cells. siRNA has recently been recognized as pivotal in the immune response of plants to virus infection. An important aspect of the siRNA response is that it normally spreads throughout the whole plant, even if the virus is contained to one tissue. Hence, detection of specific siRNAs anywhere in the plant suggests a suspected virus infection of the plant.
* One research project is focused on how to employ the plant siRNA virus response to detect known viruses and discover unknown viruses.Another research project investigates the use of cell-free smallRNAs to transfer signals in multicellular eukaryotic organisms, for instance by cancer cells.
smallRNA in eukaryotic development
smallRNAs are abundantly present in milk. This raises the question whether these smallRNAs have an immunological role in the offspring or have an antibacterial effect. With complicated, collaborative experiments, we try to address this question.
-
Research line Dr M.J. (Martijs) Jonker – Battling plant virus superspreading by humans
A critical factor in viral disease outbreaks is the distribution and spread of the hosts, vectors and viruses themselves. The recent Covid pandemic has shown that the high rate at which the virus spread was caused by unbridled travelling of humans across the globe. Similarly, to exploit favorable climate and labor conditions, man also carries large numbers of many cultivated plant species around the same globe, causing a rapid spread of plant pathogens, thus increasing the risks of disease outbreaks among agricultural crops and ornamentals. As example for recognition of this problem, this summer, European governments requested vacationers not to bring plants home to prevent the spread of the bacterium Xylella fastidiosa and in the USA government restricts firewood introduction in their national parks to limit introduction and spread of all sorts of forest pests. The risks of human induced spread of plant viruses to staple food crops such as maize, rice, wheat, potato, sweet potato, and banana have recently been recognized.
There are way more phytoviruses than vertebrate viruses, yet the focus of research and fight against viruses is clearly on the latter type. However, over the last decades, the battle between the hosts and phytoviruses tipped clearly to the viruses due to huge host reservoirs of cultivated plants plus the constant exposure to exotic viruses. This accelerated phytovirus evolution, is bad news for cultivated plants and thus the global food supply, but also for all other naturally occurring plants and thus local biodiversity.
The first step to turn the tide is to chart the problem. Fortunately, recent developments in molecular biology, such as high-throughput sequencing have made identifying new virus variants or completely new virus strains feasible. In recent years we have developed a comprehensive method (ViSiR) to identify (variants of) known phytoviruses and discover new phytoviruses by a combination of massive parallel smallRNA-sequencing of siRNAs from the plant virus defense with an advanced bioinformatics pipeline including all known virus sequences. With our ViSiR approach, we propose a project in which we, with a combination of molecular biology and artificial intelligence methods, are going to, on a large scale, identify yet unknown phytoviruses, investigate the mutation rate of plant viruses, and record the introduction of exotic virus variants in cultivated plants that are carried around the globe. Although we already employ our ViSiR approach on a small scale for Dutch plant-breeding companies, we would like to use the MMD initiative to make our approach available to all companies that use different locations around the world to cultivate their crops or ornamentals. Next to these breeding companies we see logic partnerships with controlling branch organizations, such as Naktuinbouw and NVWA.
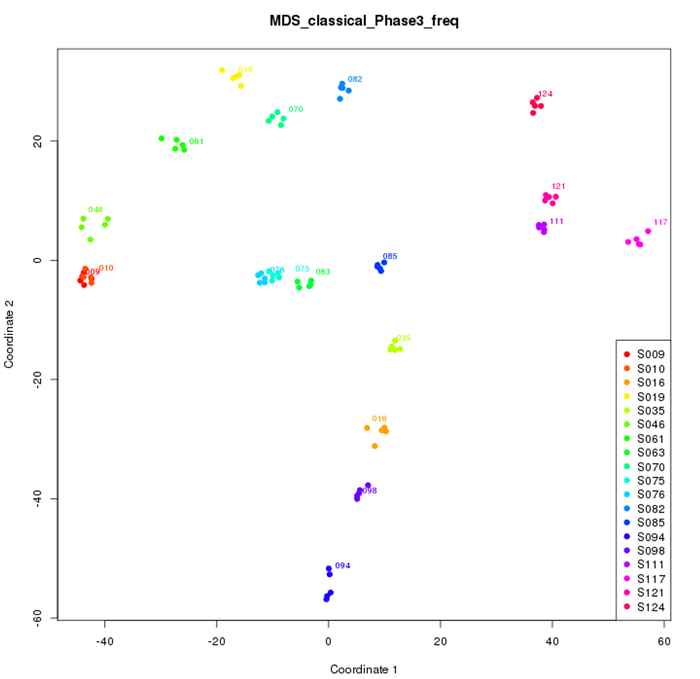
Visualizing the genomics landscape. All identified genomic differences between the analyzed accessions are used to visualize the accessions in a multidimensional scaling plot for example species X. The same colored dots are the four replicates of one accession. NB: the axes of the plot indicate the combined genomics differences between accessions. In Phase I, genomic differences in a substantial set of different accessions with replication (usually n = 4 x 24) are determined and the genomic variability between the accessions is calculated and visualized, which will result in a “genomic landscape” of the analyzed species. This phase has to be done only once for a species of interest.
In Phase II, new accessions can be placed in this species-specific genomic landscape in singular. This can be of interest for many purposes in plant breeding. For instance, one can quickly see where own or competitors’ new varieties are located in the genomics landscape.
In Phase III, we will try to focus on complex-trait biomarkers by using a genomic-prediction approach.
Contact person for this research line