Key Research Areas
Research within the Plant Hormone Biology group is focused on:
-
Rhizosphere metabolic interactions
We study the biosynthesis and the role of plant hormones and other signaling molecules in the belowground communication of plants with beneficial and harmful organisms. Additionally, we investigate how these organisms perceive these signals and the resulting impact on plant fitness.
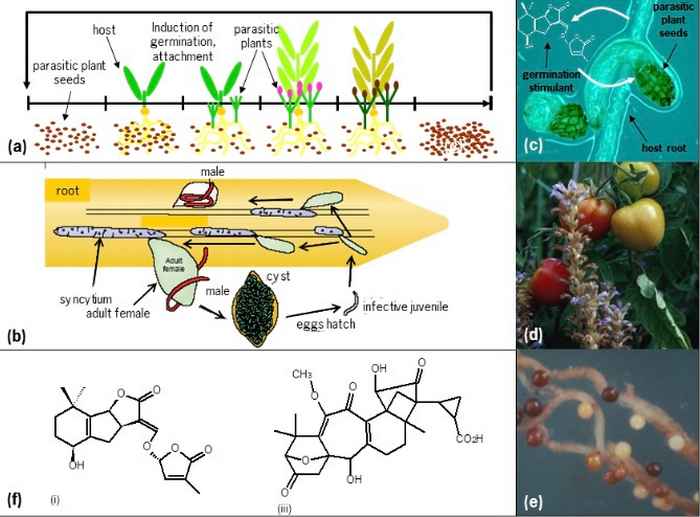
An example is the exudation of strigolactones by plants into the rhizosphere to signal host presence to symbiotic arbuscular mycorrhizal fungi and other beneficial micro-organisms. Root parasitic plants abuse this signaling relation by using the same molecules for host detection. The fact that strigolactones also have an endogenous signaling function as plant hormones, regulating root and shoot architecture, makes this interaction even more complex and intriguing.
These roles could be explained by the paradigm that was postulated first for our ERC Advanced Grant CHEMCOMRHIZO: Enemies of plants recruit molecules that are essential to the plant as cues. This paradigm has two important implications: 1) other plant-produced signalling molecules known to be abused by plant enemies likely have another, beneficial essential function in plants (as a model we study solanoeclpein A, see below) and 2) multiple, positive and negative, biological functions result in the evolution of diversity in structure and biological specificity of signalling molecules.

To investigate the latter, we study biosynthesis of strigolactones in a range of crop species. With mutants and gene editing we create plants with strigolactone profiles that differ from the wildtype to investigate what the biological relevance of individual strigolactones is. This requires further improvement of the chemical analysis of strigolactones, which we achieve through a combination of smart isolation and ultra-sensitive detection.
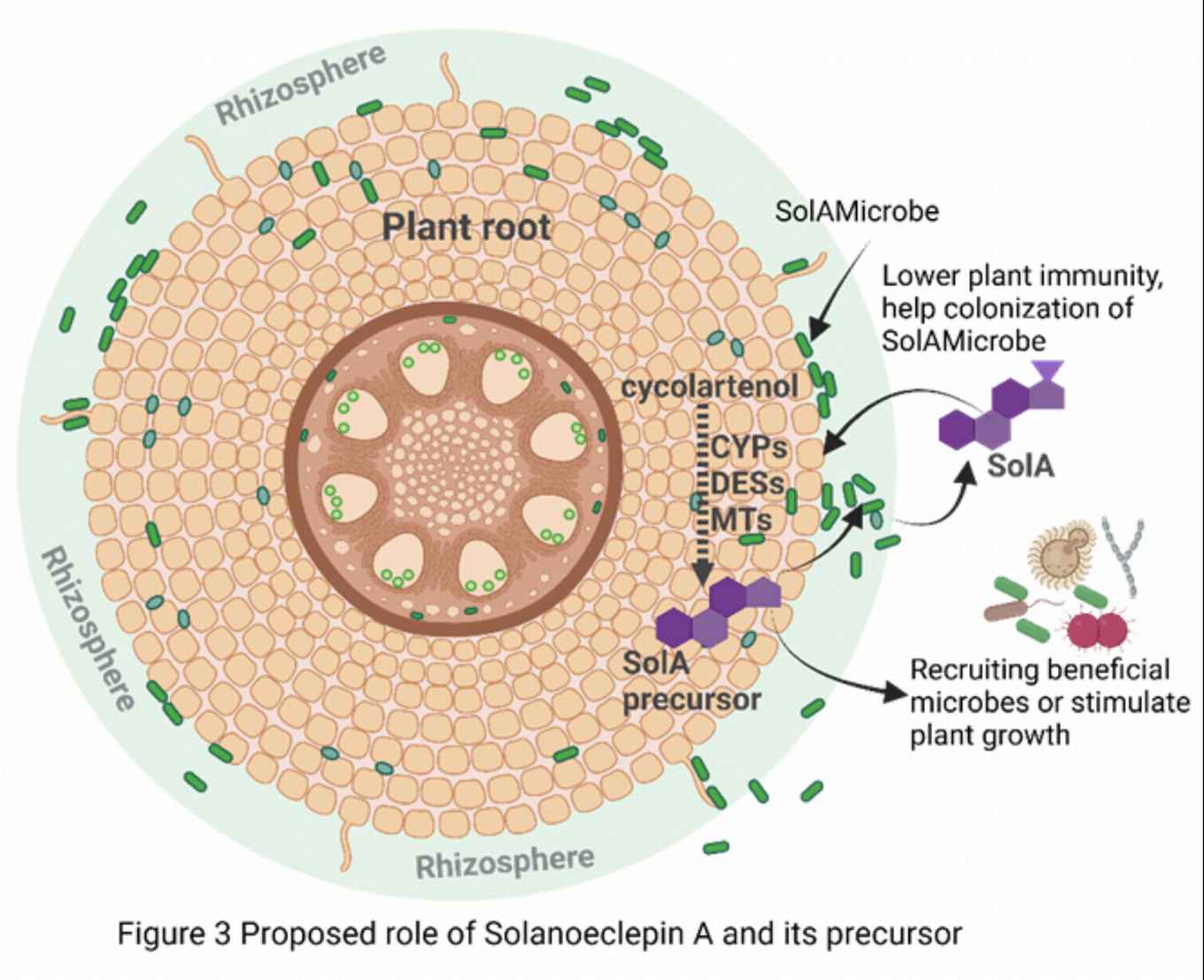
Intriguingly, in the work on solanoeclepin A, we discovered that signaling molecules secreted by plant roots can be further modified by root-associated microbes and we set out to decode these complex metabolic conversations. The model molecules we study are the triterpenoid eclepins. One of these, solanoeclepin A (SolA) was initially discovered as a host cue for parasitic cyst nematodes. Surprisingly, we unveiled that SolA constitutes a hybrid molecule requiring both plants and their root-associated microbes to be produced. We postulate that this must mean that SolA has a role other than just being a host cue for nematodes. In a number of projects funded by an EU-Marie Curie fellowship (Nemhatch), NWO-TTW, and NWO-VIDI, by combining multidisciplinary approaches in molecular biology, biochemistry and genetic engineering, we elucidate the biosynthetic pathway of SolA and other eclepins. Using omics-data analysis and microbiological approaches, we reveal the identity of the microbe(s) involved in its production (SolAMicrobe). In addition, we are uncovering the evolutionary benefit of SolA and other eclepins on plant fitness, and on colonization by microbes and nematode infection.
Our work will shed light on the significance of structural diversity in signaling molecules and may result in the discovery of new signaling molecules in plants and their further modified products in the rhizosphere. It will also contribute to our understanding of the recruitment of the root-associated microbiome and ultimately to biotechnological and agronomical applications to control parasitation by a range of organisms and to improving the interaction with beneficial organisms such as AM fungi and Plant Growth Promoting Rhizobacteria (PGPRs).
-
The microbiome for plant stress resilience and human health
The strigolactones and eclepins are very likely just the tip of the iceberg of the signalling that takes place in the rhizosphere, of which we hardly know anything due to the difficulties of studying what goes on belowground. At the same time, we know how important the rhizosphere is for plants. Especially for the root microbiome – the community of micro-organisms on and in the root of plants – it is becoming increasingly clear that it has a tremendous impact on plant growth, development and productivity and can be compared to the importance of the gut microbiome in animals and humans. To unravel relationships between plants and micro-organisms we are using targeted and non-targeted approaches. For our untargeted approach we use combinations of transcriptomics, metabolomics and metabarcoding/metagenomics.
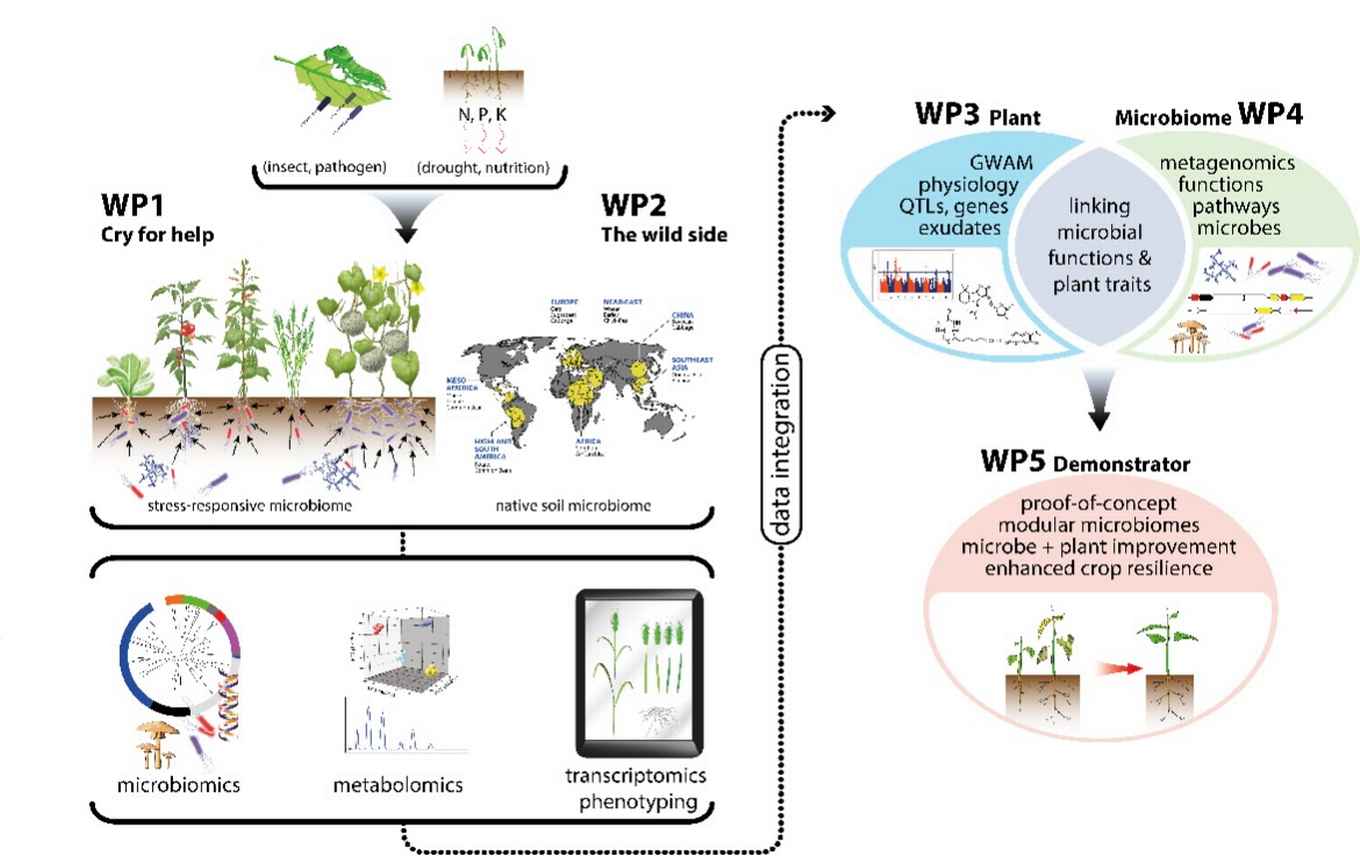
In the Gravitation project Microbial Imprinting for Crop Resilience MiCRop, together with partners in other Dutch Universities/Institutes, we investigate the role of the plant in the recruitment of beneficial micro-organisms under stress. We do this with 100 plant species across the 5 agriculturally most important plant families. In all these studies we use advanced omics approaches, data integration and modelling to try to discover new relationships between plants and beneficial micro-organisms.

We particularly focus on nutrient stress, such as phosphate and nitrogen deficiency, and the resulting signaling relationships between plants and microbes. Our research aims to uncover the phylogenetic signals conserved among cucurbit species that influence nutrient acquisition. In lettuce, we study the chemical signals that are differentially perceived by beneficial and pathogenic fungi under nutrient deficiency and examine the evolution of receptors in these fungi.

In the NWO-KIC Microbiome project, MicroHealth, we investigate the impact agriculture has on the microbiome and nutritional composition of agricultural products, and how this affects the human gut and human health. This relationship has enormous societal implications because agriculture has a large impact on our environment and on human health. With this study, we will for the first time establish the connection between the impact of agriculture on the soil and plant quality/microbiome and the consequences for the health of humans. We will also study the social and political context of this connection in collaboration with AUMC medical scientists and UvA sociologists and data scientists.

The connection with the microbiome of other organisms will be further extended in the Growthfund funded Holomicrobiome program and in which the connection between microbiomes from soil and water to food to gut is investigated.
-
Establishment of the root-associated microbiota and host preference
The diverse microbial communities inhabiting plant roots are collectively called the root microbiota. Extensive research has shown that the microbiota supports their plant host to acquire nutrients, tolerate abiotic stress, and cope with pathogen attack. Intriguingly, different plant species host distinct microbial communities, even when grown in the same soil with the same pool of micro-organisms. This suggests that there are factors specific to individual plant species that influence the attraction of and interaction with certain microbes. For example, plants communicate with their surrounding organisms through the release of signalling and organic compounds via their roots. Bacteria and fungi utilize these compounds as carbon sources for growth and proliferation but can also be either attracted or repelled from the host environment, for example by secondary metabolites and antimicrobial molecules. If we understand how the beneficial microbiota is established and maintained, we can use this knowledge to design efficient and ecologically friendly bio-inoculants as alternatives to chemical pesticides and fertilizers.
Therefore, we are assessing the following questions: Which host genes are important for microbiota assembly? How do root exudates and the microbiota affect each other? Which role do competition and cooperation within the microbial communities play for microbiota assembly and host protection?
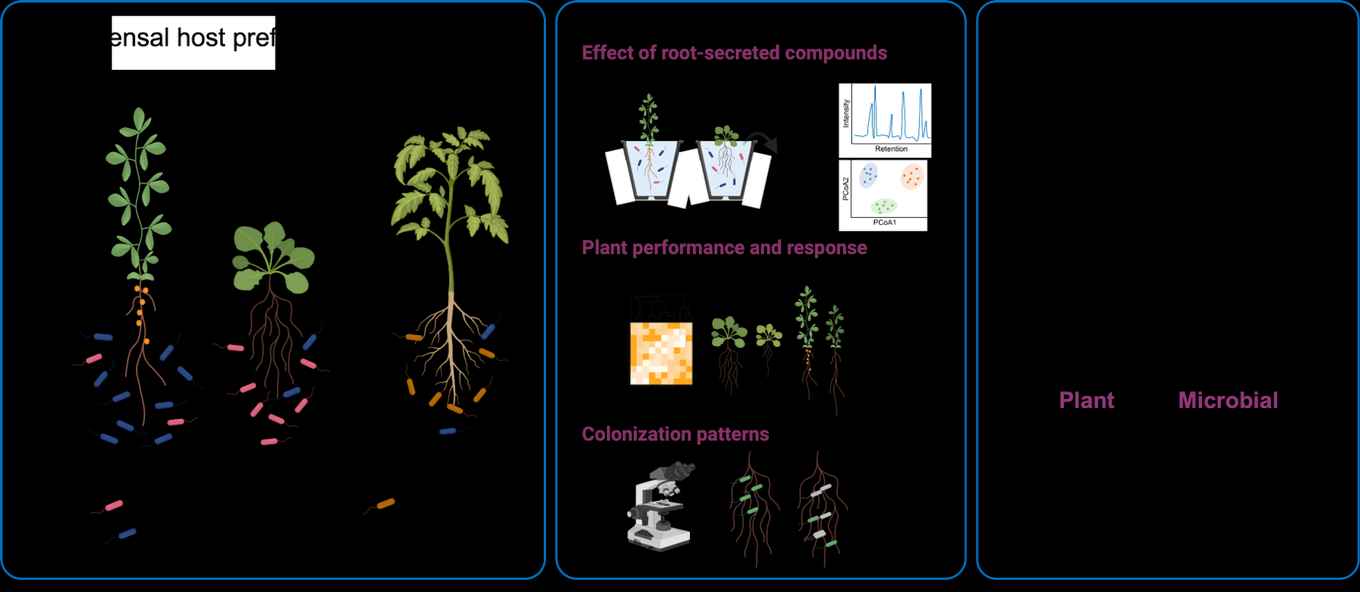
To be able to study molecular mechanisms and both host and microbial factors involved in the microbiota assembly process, we use an experimental setup of reduced complexity. We have a collection of hundreds of cultured bacterial strains isolated from different plant species, including the model plants Arabidopsis thaliana, Lotus japonicus, and the domesticated tomato cultivar Moneymaker, and whole-genome sequences for all of those strains. From this collection, we generate tailored small, synthetic communities (SynComs) and co-cultivate them with the host plants in a controlled environment. This allows us to employ plant and bacterial mutants or fluorescently tagged strains and take tissue and exudate samples, followed by several omics techniques and data integration.