Research lines Molecular Microbial Physiology
Breaking news: The Amsterdam iGEM team 2017, mentored by Filipe Branco dos Santos, has finished their project. Have a look at http://igem.jongbloets.net/
Next week, they will participate in the giant jamboree in Boston, MA, USA. You can wish them good luck on their facebook page: https://www.facebook.com/igem2017amsterdam/
The ‘Photanol approach’: using cyanobacteria to produce butanol

From Solar Energy and Carbondioxide to fuels and valuable compounds
The research of the Department of Molecular Microbial Physiology (Swammerdam Institute for Life Sciences) at the University of Amsterdam on the physiology of both phototrophic and heterotrophic bacteria is essentially fundamental. However, the combined expertise of its research leaders has resulted in a new approach to the biotechnology-based synthesis of a range of valuable organic compounds, including fuels such as ethanol and butanol. With the aid of the UvA-holding, Photanol Ltd has been established to accomodate the activities to further develop the application of microorganisms for production purposes. The company is headed by Profs Klaas Hellingwerf and Joost Teixeira de Mattos.
It is generally assumed that in the year 2050 the global annual energy consumption will have increased to at least twice the amount consumed today. Not only does this jeopardize our energy supply, but a proportional increase of the use of fossil fuels would also produce unacceptable levels of atmospheric carbon dioxide. Clearly, new technologies for the production of fuels are needed. Central to the point of view of the Photanol® concept is the fact that solar energy is plentiful (the amount that reaches the Earth's surface every hour is sufficient to meet the world's annual energy needs) but that current technologies fail to capture all that energy efficiently and store it in a useful form. Nature harnesses energy in the process of photosynthesis. In addition, Nature provides us with a broad range of biochemical pathways (called fermentations) that each produce one of a series of valuable compounds (including biofuels). Thus it is a small step to see how mankind should employ the best of these two worlds: If we would have Nature's photosynthetic specialists help us converting the greenhouse gas carbon dioxide to precursor molecules that Nature's fermentation specialists subsequently ferment to a product which we consider valuable, we have a process that contributes to lowering carbon dioxide production and exploits only the excess solar energy that is available to make valuable biofuels. This is in sharp contrast to current biobased processes that are inherently inefficient because solar energy must be invested in the synthesis of complex molecules and structures (the living plant and/or algal cells) of which only a limited set is suitable for further conversion to fuels.
Cyanobacteria, like plants, carry out the reduction of carbon dioxide (CO2) to reduced end products in a stepwise manner, as illustrated in figure 1. The intermediate products (in the figure only one such intermediate is indicated as GAP, for glyceraldehyde, 3-phosphate) formed in this metabolic pathway are the very same compounds as those used by another group of bacteria (the heterotrophs) that carry out fermentation (figure 2). The Photanol® concept merges the biochemical properties of these two groups of bacteria by constructing a cyanobacterium that carries out photosynthesis and contains the gene cassette needed to convert GAP to whatever desired fermentation product, e.g. ethanol, propanol, butanol, etc. etc. The overall scheme is presented in figure 3.
The overall equation of the chemical reaction catalysed by the photofermenting organisms then reads:
Solar energy + CO2 + H2O -----> fuel (or any other desired product) + O2
This illustrates the essence of the Photanol concept: a clean, carbon dioxide consuming, highly efficient and flexible production platform for a wide variety of fuels and chemicals.
People working on this Research line:
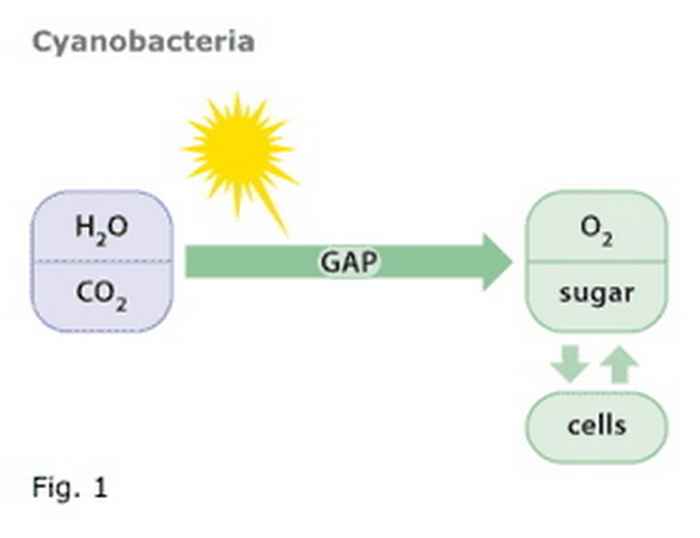
Figure 1
Cyanobacteria bacteria use solar energy to oxidize water to oxygen and to reduce CO2 to sugar. From sugar new cells are made. GAP is an intermediate in this biochemical pathway.
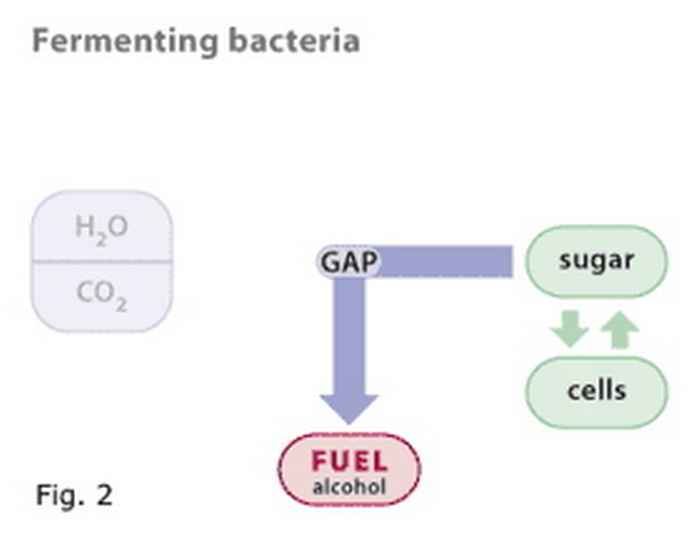
Figure 2
Fermenting bacteria use sugar to make new cells, and as their energy source. The end product of the energy yielding biochemical pathway may be any of a variety of organic compounds, including alcohols. Again, GAP is an intermediate.
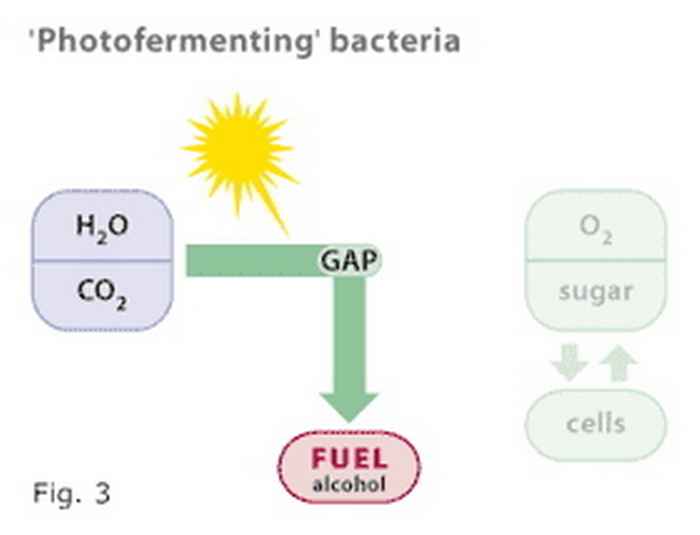
Figure 3
The Photanol concept combines the properties of photosynthetic and fermenting bacteria into a 'photofermenting' bacterium that uses solar energy to reduce CO2 to any of a variety of organic compounds, including alcohols. Oxygen is the only waste product.
Comparative Systems Biology: Lactic Acid Bacteria
This project focuses on three relatively simple and highly related microorganisms, which nevertheless exhibit stark and important differences in their functional relationship with human beings.
These organisms are homofermentative lactic acid bacteria, namely:
Lactococcus lactis, the major microorganism used in the dairy industry,
Enterococcus faecalis, a major (faecal) contaminant in food and water as well as a contributor to food fermentation, and
Streptococcus pyogenes, an important human pathogen.
These organisms have a similar primary metabolism, but persist in completely different environments (milk, faeces, blood).
Lactococcus lactis will be used here as the reference microorganism, since:
(i) it is by far the best studied lactic acid bacterium,
(ii) three different genomes have been sequenced,
(iii) a kinetic model has been developed for its complete glycolysis including some branching pathways.
Comparative analyses is used for transfer of information from the (experimentally) well-studied organism Lactococcus lactis to the organisms E. Faecalis and S. pyogenes. Functional and phenotypic information will be used to gain insight in the relative importance of components (e.g. metabolite concentrations, Km or Vmax of a specific enzyme) to the observed differences and similarities (e.g. glycolytic flux).
The central principle of this project is that important aspects of the functional differences between organisms derive not only from the differences in genetic components (which underlie comparative genomics) but also from the interactions between their components (e.g. interactions between enzymes, feedback loops etc). This multidisciplinary approach will result in mathematical, comprehensive models that quantitatively describe the catabolic (product formation) response of lactic acid bacteria under different conditions. These models will help in the design of strategies that optimise functional activities such as acidification and flavour production (for the industry), but also strategies to control or reduce growth of undesirable contaminants and pathogens.
People working on this Research line:
Proteomics studies of Escherichia coli and Synechocystis sp. PCC 6803
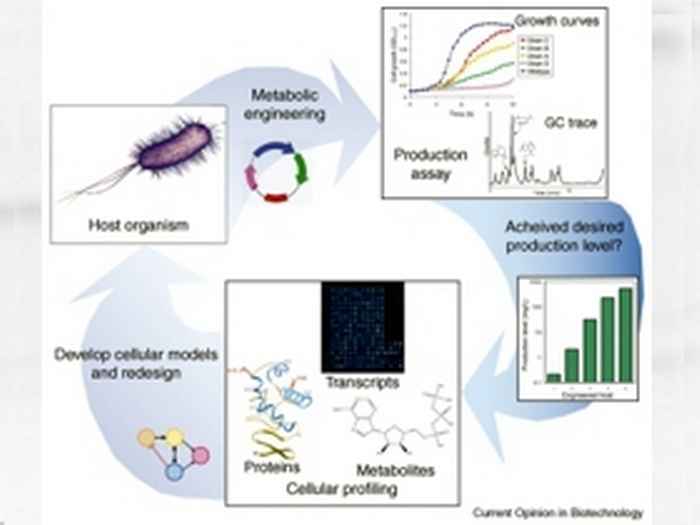
Systems biology:
Current research in molecular physiology of microorganisms is characterized by the deployment of 'omics' techniques, to provide systems-wide information about (the regulation of) basic cellular processes such as transcription, translation and intermediary metabolism. The information thus obtained then allows one to set up detailed mathematical models of cellular systems. With such models an iterative process can be initiated in which gradually cellular understanding can be brought to the molecular level of all aspects of a living cell: The systems biology approach.
The most straightforward of these omic technologies is based on the analysis of transcript profiles, with the help of oligonucleotide micro-arrays. This technique by now is well-established, to the extent that generally accepted quality guidelines have been accepted by the research community with respect to the type of experimental protocol to be used for such experiments. This status was achievable because of the completeness and non-ambiguity with which genome sequence data can be obtained, in combination with well-developed protocols for the isolation of cellular RNA and oligonucleotide hybridization. A similar situation is not yet within reach for the two complementary omics techniques, proteomics and metabolomics. The much larger variety in chemical structure of the classes of molecules to be analyzed with these techniques significantly complicates their comprehensive analysis. In this project we will further develop the tool box for microbial proteomics analysis.
Organisms and processes to be investigated:
Our studies will focus on two separate processes that are of basic interest in molecular microbial physiology:
(i) Catabolite repression in the model organism Escherichia coli
(ii) Proteomics analysis of biofuel-producing Synechocystis sp. PCC6803 strains.
In both organisms, proteomics analyses will be carried out in dynamic and steady state conditions, using various approaches such as ITRAC, SILAC, etc., with emphasis on both soluble and integral membrane proteins, thereby extending earlier work (see; PubMed Kramer et al and Nessen et al).
People working on this Research line:
On the nature of diffusive transport in photo-activated cell signaling in baker’s yeast
On the nature of diffusive transport in photo-activated cell signaling in baker’s yeast.
The focus of this project is on the operation of biochemical regulation networks at the cellular level and in particular the spatial design principles of such networks. Itsfour main goals are:
- Construct a new light-sensitive kinase by fusing a kinase domain from Saccharomyces cerevisiae with a photoreceptor domain from Bacillus subtilis.
- Measure cytoplasmic/nuclear shuttling of a GFP-labeled reporter protein in response to cell signaling activated by periodic light stimulation of the cell globally and locally.
- Determine amplitude and phase response of the diffusive response as a function of activation frequency.
- Study whether compartmentalization affects and/or limits cellular responses.
Saccharomyces cerevisiae Sln1p is a histidine kinase belonging to the family of two-component regulation systems, which play a prominent role in the response of prokaryotic organism to the extracellular environment. Sln1p, which is functionally active as a dimer, auto-phosphorylates one of its conserved histidines in response to changes in the osmotic pressure in the environment. This phosphoryl group is then transferred in trans to a conserved aspartate in the Sln1 receiver domain, subsequently to a histidine in the phosphoryl-transfer domain Ypd1, and finally to an aspartate residue on the receiver domains of the response regulators Ssk1p and Skn7p
A promising route to be able to determine the role of compartmentalization and diffusion in signal transmission in the eukaryotic cytoplasm is provided by the photo activation method. For this we plan to make use of YtvA from Bacillus subtilis. This is a photoreceptor protein, comprised of an N-terminal LOV sensor domain and a C-terminal STAS (sulfate transport antisigma factor antagonist) effector domain, joined by an alfa-helical linker sequence denoted Jalfa. It can elicit the general stress response in B. subtilis upon blue-light absorption. In the dark, its LOV domain binds flavin mononucleotide (FMN) non-covalently. Absorption of blue light promotes formation of covalent bond between the flavin ring and the conserved cysteine 62 of the LOV domain.
In Saccharomyces cerevisiae strains deleted for Sln1p we will engineer a light-dependent soluble histidine kinase, by combining the LOV domain of YtvA and the kinase domain of Sln1p. This fusion protein will then be used to phosphorylate GFP-Labeled Ypd1p in a light-dependent manner, which then will accumulate in the nucleus.
This research is a part of the project “Spatial design of biochemical regulation network”. This program brings together biophysical and biological expertise in the experimental study of gene and protein networks, the (spatial) modeling of biochemical regulation networks, and the spatial organization of cytoskeletal polymer networks that are responsible for active linear transport of regulatory proteins in cells.
People working on this Research line:
Population heterogeneity in a probiotic lactic acid bacterium
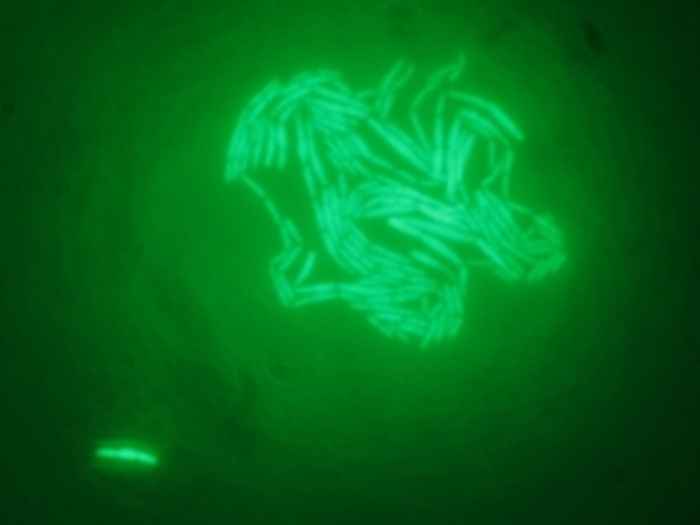
During production, downstream processing and storage of probiotic dairy products, the probiotic culture gradually loses viability due to a combination of various stresses, such as oxidative stress, temperature stress and starvation. To maintain minimally required viable counts in the probiotic products over extended periods of time, it is essential to identify microbial strategies that lead to increased survival
Biological parameters of bacterial strains, such as growth rate, metabolite production and response to environmental stress are determined in cultures that consist of billions of single cells. This average is justifiable as long as the individual parameters of cells are normally distributed. However, when a subpopulation of cells is present that shows deviant behavior to the majority of cells, this indicates that the diversity is not merely the result of stochastic differences, but has an underlying mechanism. For example, when continuously small subfractions of cells were found that showed multi-drug tolerance (so-called persisters), it was discovered that this resistance is a property of a subpopulation of non-growing cells.
In this project, the hypothesis that population heterogeneity plays a role in survival will be tested for a probiotic lactic acid bacterium. Inactivation experiments suggest that sub-populations occur that show variable resistance against environmental stress. The existance of such subpopulations can be observed by counting colony forming units on agar plates, by following microcolony formation on Anapore micro-slides and by Fluorescence Activated Cell Sorting (FACS) using live-dead staining.
People working on this Research line:
Structure/function relations of photosensory- and fluorescent proteins

Both photoreceptor and fluorescent proteins enter a photocycle after absorption of light. Light is an important part of the environment of an organism. For surviving, organisms have to respond and adapt to (changes in) their environment. Light is sensed by photoreceptor proteins. Photoreceptor proteins are adapted to absorb light of particular wavelengths and transmit the signals to response regulators. Fluorescent proteins such as GFP absorb light of particular wavelengths and emit light with a different wavelength. The characteristics of a protein are determined by its amino acids. Not all amino acids in a protein have the same degree of importance. Some amino acids can be replaced by another without it having an effect on the characteristics of the protein, while mutagenesis of other amino acids can lead to inactivation of the protein. In this project the photocycle of several photoreceptors and fluorescent proteins are studied by site-directed mutagenesis. Most intermediates in the photocycle can be distinguish by there absorption spectrum. Therefore, the photocycle of isolated proteins can be followed in time by UV/Vis spectroscopy. Also fluorescence spectroscopy is used for this study.
Photoactive Yellow Protein (PYP)
PYP is a prototype of a PAS domain. The small size (125 amino acids) of PYP and its high water solubility make it an easy protein to work with. The most studied PYP is from the purple sulphur bacterium Halorhodospira halophila. After absorption of blue light, PYP enters a photocycle in which the double bond of the chromophore isomerizes. Via several intermediates, the signaling state is formed. In the signaling state the protein is partially unfolded. The photocycle is completed by recovery back to the ground state. In seconds after a light flash PYP goes through its photocycle and is back in the ground state.
BLUF domain of AppA
AppA from the facultatively photosynthetic purple bacterium Rhodobacter sphaeroides regulates photosynthesis in response to changing oxygen tension and light intensities. AppA contains two domains, a C-terminal domain for redox sensing and an N-terminal BLUF (blue-light using flavin) domain. The signaling state of the BLUF domain is formed in 1 ns and has a relatively long half-life of ~15 minutes. In the signaling state, the FAD has the same chemical state as in the ground state. Only the hydrogen bonding network around the chromophore is different. AppA is studied in vitro and also in vivo.
Green fluorescent protein (GFP)
GFP has a remarkable bright green fluorescence. The most studied GFP is from the jellyfish Aequorea victoria and has become a popular fluorescent marker. After absorption of UV or blue light, it emits green light. GFP follows a complex photocycle in which a proton transfer takes place. The chromophore of GFP is formed from the three amino acid Ser65, Tyr66, and Gly67.
Unraveling mechanisms by which environmental factors affect catabolic efficiency
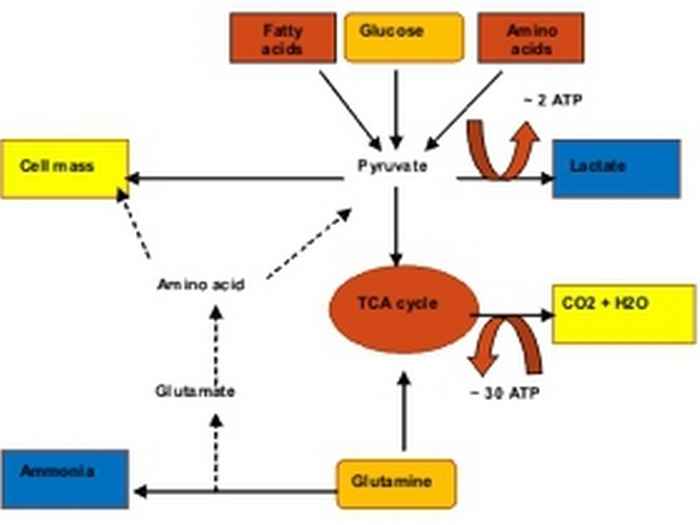
Unravelling environmental factors that affect catabolic and anabolic efficiency
The interest in the application of various mammalian cell types for tissue engineering, for the production of proteins (e.g. antibodies) and for the manufacturing of artificial meat has increased now that large scale culturing technologies have become available. Further optimization of mammalian cell-based biotechnology demands quantitative insight in a) the metabolic machinery of these cells b) its regulation and c) the energetics of growth and maintenance. This project aims at a quantitative framework that describes these issues and thus to provide the tools to optimize production parameters like growth rate, growth prolongation, product formation and nutritional demands. A typical example of biotechnologically relevant cells are Chinese Hamster Ovary (CHO) cells which have gained an important industrial interest as successful platform for the biotechnological production of numerous valuable compounds. Although proven to be applicable, there is a great demand - and ample room - for optimization of production processes with respect to volumetric- and specific production rates on the one hand, and prolongation of production on the other.
The low energetic efficiency of CHO cells when grown on glucose as the energy source, is due to the glucose flux being directed towards fermentation, yielding lactic acid as end product. Although in many cases it may be due to insufficient aeration, also properly oxygenated cells express high glycolytic rates on the one hand, and low oxidative activity of the tricarboxylic acid (TCA) cycle and low respiratory activity on the other. In addition, it should be realized that the in vivo concentrations of oxygen are usually much lower than those to which cultured cells are exposed. This lower efficiency of catabolism not only has adverse effects on the yield value (no. of cells per mole of glucose), but will also affect growth rate, longevity and volumetric productivity in a negative manner.
The growth rate, number of generations (longevity) and metabolic rates are all dependent on environmental conditions. A quantitative assessment of the specific uptake and conversion rates of the carbon- and energy sources such as sugars, peptides, amino acids and fatty acids is paramount for an understanding of the metabolic network of these cells. Most importantly, the availability and the nature of the energy source (i.e. glucose, fructose, lactose), relative to the amino acids (specifically glutamine) is thought to play a role in the rates of sugar transport, TCA cycle activity and ammonium production. Mammalian cells are able to sense and respond to the extracellular glucose level by regulators like PPARs and these may play a role in glycolytic activity as well, and possibly even more downstream parts of catabolism. Therefore they seem to be proper targets to modulate the efficiency of energy conservation.
Production efficiency is highly dependent on the metabolic activity of the cell, including both the specific production rate (production per cell) and the biomass density (which is dependent on the cell's efficiency of energy conservation) that can be obtained under a given growth condition. The generated knowledge should lead to a hypothesis as to how production of some heterologous protein be optimized. However, it should be taken into account that diverting anabolic fluxes from cell synthesis to (extracellular) protein synthesis may have multiple effects on intracellular pools (of amino acids and their precursors, e.g. pyruvate). As a consequence, catabolic gene expression, related enzyme activities and energetic demands may alter in an unforeseen way.
Current interest in the culturing of (muscle derived stem) cells for the purpose of manufacturing artificial meat (additives) has initiated research into the design of cheap media for large scale production of eukaryotic cells. These cells are dependent on growth factors and other, often undefined, complex nutrients for growth (and proliferation) . Rather than using serum (e.g. foetal calf serum) as the source for these nutritional compounds, it seems feasible to have the recombinant growth factors produced by engineered strains and other commercial hydrolysates with the necessary simple salts (phosphate, potassium, calcium, etc), and a carbon and energy source.
People working on this Research line:
Studies of cellulose synthesis in cyanobacteria

Similar to the Photanol® approach (see: top of this page), this project aims to convert the carbon products of phototrophic organisms, i.e. glyceraldehyde-3-phosphate (GAP), into products that can be harvested efficiently in terms of economy and yield. Different approaches have been proposed to achieve this. One is to harvest biomass and convert it thermo-chemically into syngas. Another is to grow photosynthetic organisms with high triglyceride content, followed by conversion of this lipid fraction, via trans-esterification, into biodiesel. A third approach aims at direct conversion of CO2 into biofuel (Photanol approach). Yet these approaches suffer from problems in the down-stream processing of the ‘fixed-carbon’ product.
In this project we explore optimization of downstream processing of a fixed carbon product. We aim at accumulation of the primary product of carbon fixation in photosynthesis at high yield, more than 90% of dry weight, in an extra-cellular polysaccharide (EPS, i.e. cellulose). Cellulose is optimally suited to be converted to biofuel. Although cellulose can be amply harvested from trees the extraction of cellulose from wood requires harmful chemicals due to the presence of non-degradable compounds such as lignin. And when using cellulose from cyanobacteria no forest will have to be cut down.
Several cyanobacterial strains, including Crinalium epipsammum, have been demonstrated to naturally synthesize high amounts (20% of dry weight) of crystalline cellulose. It is expected that this yield can be considerably increased through manipulating culture conditions. Furthermore we will use a synthetic biology approach to engineer a strain of the cyanobacterium Synechocystis sp. PCC 6803 with high cellulose yield by the addition of a crystalline cellulose extracellular accumulation system.
By applying rapid quench- and extraction techniques the composition and redox state of the quinone pool(s) of these Synechocystis cells will be monitored. This will provide key information for future work to make quantitative models of cyanobacterial primary metabolism relevant for the photanol process.
People working on this Research line:
Characterization of a large signaling complex in the general stress response of Bacillus subtilis
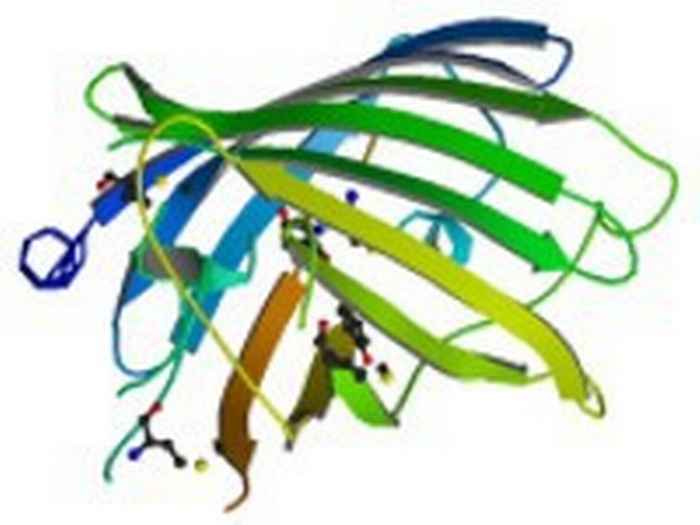
Bacillus subtilis is a bacterium commonly found in nature, in for example soil and water, and in association with plants. It is an interesting organism for a number of reasons.
For one, Bacillus subtilis is the second-most important cause of the spoilage of bread (after fungi). Several other members of the Bacillus genus can also spoil food, and some can even cause food poisoning or disease. Perhaps the best-known example of the latter is Bacillus anthracis, which causes the disease anthrax.
However, Bacillus subtilis and its family members can also be used to our advantage. Examples include its use as a fungicide or a human and animal probiotic, but Bacillus subtilis is also one of the most widely used organisms for the industrial production of enzymes, and there are several additional fields in which its potential usefulness has not yet fully been exploited.
The General Stress Response
The natural environment of Bacillus subtilis is prone to sudden changes, which can be very unfavorable for the bacterium. To counteract these 'stresses', B. subtilis has developed a wide range of specific responses.
Additionally, the organism has developed a general stress response (GSR) which responds to a wide range of stresses and provides relatively low-level protection.
These stresses include nutrient deprivation (= energy stress) and salt-, ethanol-, acid- and heat-shocks (referred to as environmental stress).
When activated, the GSR modulates the expression of about 150-200 genes involved in, amongst others, specific and non-specific stress protection. Mutants lacking a functional GSR usually have problems surviving any of the aforementioned stresses, which clearly shows the importance of this protective mechanism.
The GSR is regulated by a complex protein network which goes beyond the scope of this introduction. An intriguing part of this network is a large protein complex called the stressosome.
A number of regulatory proteins is involved in the stressosome, including (surprisingly) a blue-light photoreceptor protein, which can activate the GSR when it is illuminated.
Project
In this PhD project we aim to resolve the molecular basis of the structure and functioning of the stressosome. The aspects we will address include, but are not limited to: the localization of the stressosome using GFP-fusions, its interaction with the aforementioned blue-light photoreceptor, the identification and mode of action of the signals that make the stressosome respond to stresses (including light), and the construction of a mathematical model to describe the interactions in this complex regulatory network.
People working on this Research line:
Understanding the respiratory chain of Escherichia coli - How do bacteria breathe?
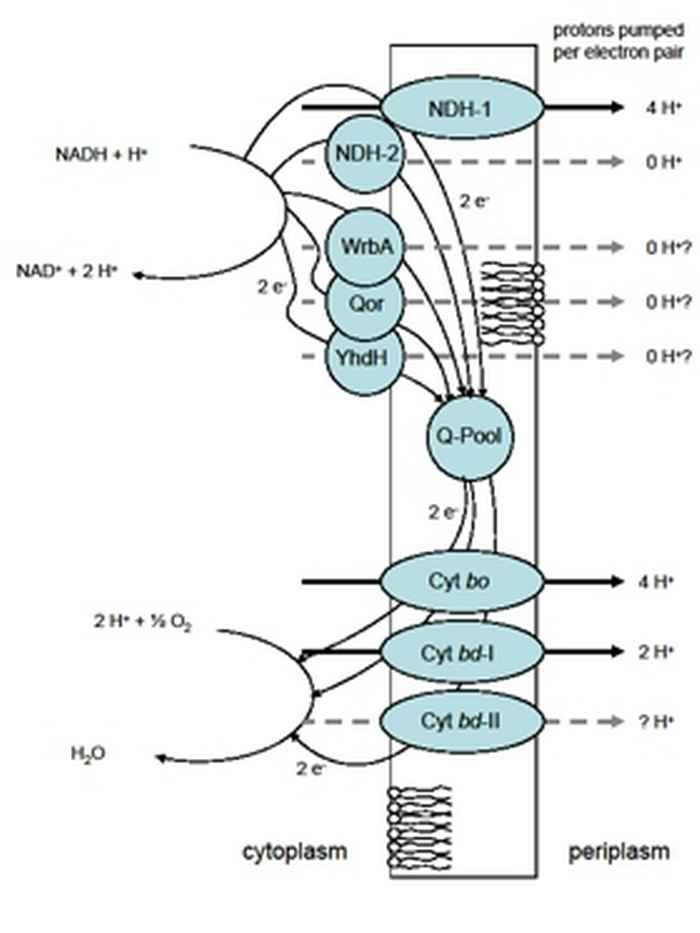
Oxygen is vital for many organisms. However, many bacteria are able to proliferate under low or no oxygen conditions. The Gram-negative bacterium Escherichia coli grows most efficiently under aerobic conditions, but is also able to reproduce under micro- and even an-aerobic conditions. This micro-organism is commonly found in the lower intestine of warm-blooded animals. Most Escherichia coli strains are harmless, but some can cause serious food poisoning in humans. Further, this bacterium is widely used in all kinds of biotechnological applications e.g. for the industrial production of chemicals of therapeutic or commercial interest
In the respiratory chain, electrons flow from the electron donor NADH (generated in carbon catabolism) via the quinone-pool to the final electron acceptor oxygen. The electron flow is catalyzed by a number of NADH dehydrogenases and cytochrome oxidases. Since the NADH dehydrogenases function in parallel and the same is true for the oxidases, the electrons can flow from NADH to oxygen through different routes: the respiratory chain of Escherichia coli is branched. The redox energy that becomes available when electrons flow from NADH to oxygen may be converted into a proton gradient over the membrane. This gradient can subsequently be used to make ATP (the main energy storage and transfer molecule in the cell) via a membrane-bound ATPase. The efficiency of energy conservation is dependent on the distribution of the electron flow over the alternative dehydrogenases and oxidases. Thus, Escherichia coli has a respiratory chain that is highly flexible and readily adaptable to the energetic needs of the cell.
The main goal of this project is to understand the branched respiratory chain of Escherichia coli. We are trying to answer the following questions: -Why is the electron transfer chain of Escherichia coli branched? -Under which environmental conditions (oxygen availability) are the different respiratory chain components needed? -How is the flow of electrons distributed over the different components in the respiratory chain under the different conditions? This is done by growing the Escherichia coli in well controlled chemostats, in which we can set the oxygen availability and other environmental parameters. Besides the wild-type Escherichia coli strain, many specific (linear) respiratory mutant strains are used to control the (proton translocating) efficiency of the respiratory chain. This allows us to describe the catabolism of the cell in terms of specific fluxes (glucose and oxygen consumption, product formation and growth rate) and to calculate the energetics of microbial growth.
This research is part of a trans-national project called Systems Understanding of Microbial Oxygen Responses (SUMO). The aim of the consortium is to understand and to model the oxygen response of Escherichia coli by a systems biology approach. Deeper understanding of fundamental processes like respiration is of general scientific interest and in addition will help to improve industrial fermentations and process scale-up. Further, the availability of oxygen plays a role in the virulence of many pathogens. Hence, our research may contribute to fight these organisms. SUMO is part of the Systems Biology of Microorganisms (SysMO) initiative that is funded by several European funding agencies (www.sysmo.net).