UvA biologists find new method of DNA organization in bacteria
31 January 2022
Bacteria are relatively simple if you compare them to organisms such as plants, animals and fungi. One big difference is that the cells of all the latter organisms contain a nucleus, which holds the cell’s DNA; whereas the cells of bacteria lack a nucleus. Their DNA floats freely within the cell. Biologists call organisms which cell’s have a nucleus “eukaryotes” and those without a nucleus “prokaryotes”. There are of course still similarities between the two. For both, DNA is the genetic code that serves as a blueprint to make the cells function. Also, in eukaryotes as well as in prokaryotes the strands of DNA are so long that they need to be folded into complicated structures to fit.
DNA loops and genes
We know that the precise way in which these DNA structures are organized play a role in what parts of the genetic code can be read or not. DNA contains genes, that can be either switched “on” or “off”. The most direct way to switch on a gene is via a protein that binds on the DNA in the direct vicinity of the gene. But in eukaryotes, more complex mechanisms are found where a protein binding to a part of the DNA far away from the gene can still switch it on if the DNA is folded in such a way that the two regions get looped closely together. In prokaryotes, such DNA looping proteins (or anchors) which connect DNA regions that are far apart remained elusive. Until now.
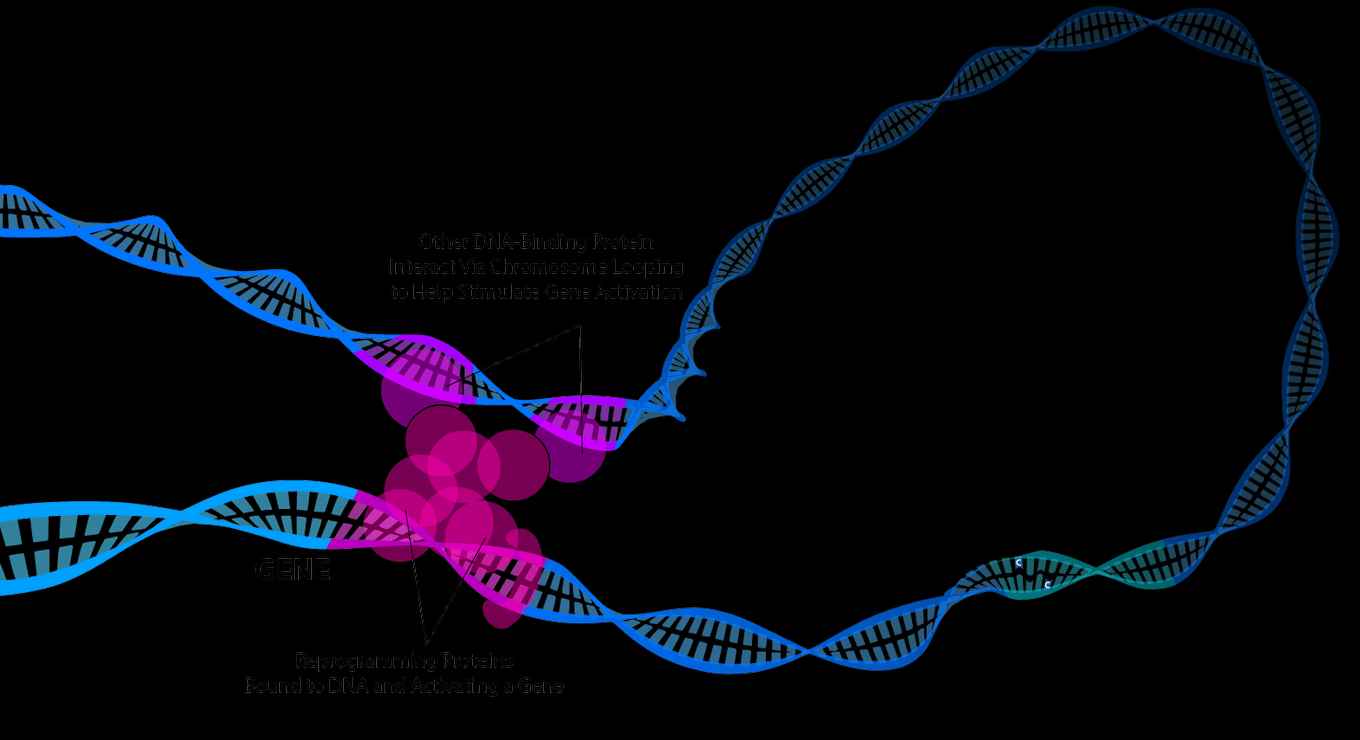
Proof in bacteria
Dugar and Hamoen teamed up with colleagues from the Heidelberg University in Germany. Together they found proof that a protein named Rok can connect DNA regions that are far apart in order to switch gene expression in the soil bacteria Bacillus subtilis. The researchers managed to map the interactions of the whole DNA using a global DNA capture technique. They were able to visualize the formation of DNA loops connecting regions of the DNA separated by millions of nucleotides. This is somewhat similar to the DNA loops already known in eukaryotes for decades. Furthermore, the researchers found that the genes in these DNA loops were spatially isolated from the rest of the genome. This is the first study to show that such DNA interactions do exist and affect gene regulation in bacteria.
'Like most bacteria, Bacillus subtilis DNA is composed of a single large circular chromosome.’, comments Dugar. ‘We were quite surprised to see that several sections of the chromosome disappeared when we separated it by density. Later, we found that these DNA sections were associated with the protein Rok, which connected these sections together to make large DNA loops within the chromosome. This was quite exciting as such DNA loops were only observed within the nucleus of eukaryotic cells previously'.
Outlook
While previously biologists thought that bacteria had only direct mechanisms for regulating and reading the genetic code, this study shows that it is more complicated. The regulation and folding of DNA adds an extra layer to how genes can be regulated and how cells can react to their environment. With the methods set up by Hamoen and colleagues, more candidate proteins can be investigated to help unravel more about the diversity and functionality of this process that is fundamental to the organization of bacteria, and all kind of cells and life in general.
The study also shows that even though we now know more about genetics than ever before, there is still a lot out there waiting to be discovered even in seemingly simple organisms. Implications of these findings cannot be caught in direct terms yet. Knowing more about such a fundamental process opens the way towards a better understanding of DNA folding and regulation in general, and this knowledge can be helpful in all kinds of ways, for example to target harmful bacteria in a way previously not known. But for now, it mainly means that much is still to be discovered about this interesting mechanism and its role in bacteria.
Details of the publication
Dugar, G., Hofmann, A., Heermann, D.W. et al. A chromosomal loop anchor mediates bacterial genome organization. Nat Genetics (2022). https://doi.org/10.1038/s41588-021-00988-8
You can find more information about the Research Group 'Bacterial Cell Biology' here