Dr. ing. F.L.W. (Frank) Takken
- Science Park 904
- Room number: C2.214
-
Postbus 1210
1000 BE Amsterdam
-
Research
Research
Since 2001, my group studies the molecular and biochemical mechanisms underlying disease -resistance and -susceptibility in plants. We want to understand how the immune system of a plant allows it to recognise a pathogen and mount effective defence responses. Furthermore, we want to elucidate how pathogens manipulate their hosts and manage to evade its immune system.
As model crop, we use tomato because of its great genetic tools and its importance for Dutch agriculture. Tomato has many natural enemies and one of them is the root-invading soil-borne pathogen Fusarium oxysporum, the causal agent of Fusarium wilt disease. Resistance to fusarium can be conferred by Resistance genes. These genes encode immune receptors that recognize specific proteins secreted by the fungus. Our aim is to understand how NLR-type immune receptors function in disease resistance. Biochemically we have shown that these proteins act as nucleotide-operated molecular switches. In collaboration with the Cann laboratory (Durham UK) we found that these proteins can bind and bend DNA in vivo, and that in planta they bind DNA once activated. In an NWO funded VICI project we studied the role of DNA binding by NLR proteins in plant immunity. Using our knowledge of immune receptors we now aim to adjust their recognition specificity to allow them to recognise newly emerging pathogens. For this reserach we focus on NLR receptors from solanacea recognising fungal, viral and bacterial pathogens.
Tomato is one of the model crops used in our lab. One of the main advantages is that is is easy to make crosses Although Fusarium oxysporum is infamous for being a devastating pathogen, many strains are actually endophytes that enhance plant resilience to biotic and abiotic stresses. Current research aims at resolving how endophytes evade host resistance while boosting plant immunity, and to elucidate how endophytes differ from pathogens.
Disease resistance can also be conferred by a lack of compatibility with a given pathogen. To unravel this mechanism, we focus on the identification of compatibility/susceptibility genes of the host by screening for protein targets that are manipulated by a pathogen. Upon infection Fusarium oxysporum secretes many small proteins in the xylem sap of the host. Many of these Six (Secreted In Xylem) proteins, such as Six3, Six5 and Six6, are important for disease development, like. We use these Six proteins to identify their plant targets and identified genes whose knockout result in increased disease resistance. Identification of these susceptibility genes increases our understanding on how a pathogen manipulates its host and providing new leads to combat diseases
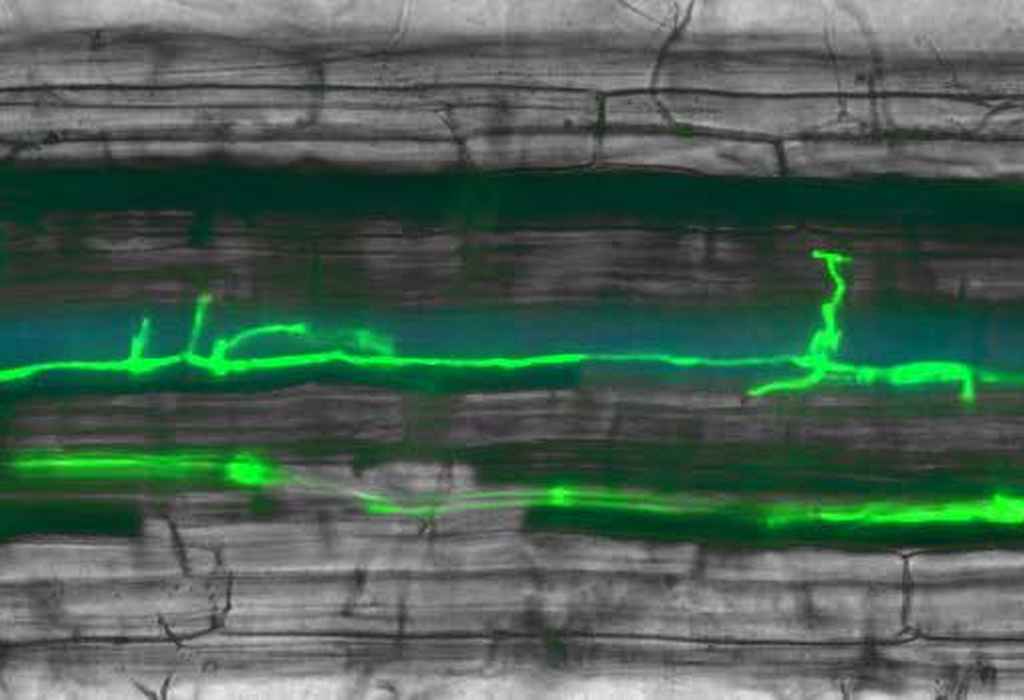
Besides tomato, we use Nicotiana benthamiana to transiently express genes and to study the action and location of recombinant proteins. In this leaf the I-2 Resistance gene is co-expressed with a fungal Avr2 genes. A hypersensitive cell death response is induced where both proteins are present. Agroinfiltration in N. benthamiana. -
Research Team
Group members
Current members of the research team.
Machiel Cligge PhD student - "Virus induced genome editing in crops to facilitate precision breeding", TKI Topsector AGri& Food
Margarita Simkovicova PhD student - "Combating Vascular Diseases: Identifying the Guardians of the Xylem Sap", Collaboration with ENZA zaden, funding by ENZA and NWO-VICI program
Thomas Aalders PhD Student- TOPLESS a novel S gene: Unravelling the manipulation of Topless by a conserved effector from Fusarium oxysporum", funding by NWO talent program:
Dr Daniel Zendler Postdoc - “Integrated research program towards sustainable resistance against the Tospovirus-thrips pathosystem in crops”. Topsector T&U project LWV20.105
Babette Vlieger PhD student -" A leap into the unknown: understanding host-jumping by Fusarium oxysporum in cucurbits" NWO talent program
Jo Gomila technician -"Precision genome editing to alter disease resistance specificities in plants" TKI Topsector AGri& Food
Dr Isidre 'd Hooghvorst - -"Precision genome editing to alter disease resistance specificities in plants" TKI Topsector AGri& Food
Tomato transformation and regeneration. Former lab members
Dr Mila Blekemolen, PhD student - NWO VICI program and UvA: Elucidating the molecular mechanism underlying the virulence and avirulence functions of the Fusarium Avr2/Six5 effector pair
Dr Nico Tintor Postdoc - ENZA project: effector-uptake mediated plant immunity
Dr Marijn Knip, Postdoc - VICI project: Linking plant immunity and DNA damage.
Dr Manon Richard, Postdoc - VICI project: Linking DNA binding of plant NLR proteins to plant immunity.
Machiel Beijaert, Technician - VICI project: supporting the VICI project in various ways.
Dr Francisco de Lamo Ruiz, PhD student - ETN Horizon2020 : How do endophytes evade host resistance while enhancing immunity?
Dr Maria Constantin, PhD student - ETN Horizon202: How do endophytes differ from pathogens?
Dr Lingxue Cao, PhD student - CSC grant: Elucidating the molecular mechanism underlying the virulence and avirulence functions of the Fusarium Avr2/Six5 effector pair,
Dr Xiaotang Di, PhD student - CSC grant : The Avr2 effector protein from Fusarium as a guide to unravel plant innate immunity.
Dirk-Jan Valkenburg, Technician: involved in various ongoing projects in the group.
Dr Hanna Richter, Technician: involved in all ongoing projects in the group.
ir. Lisong Ma, PhD student: Analysing the intrinsic functions of Avr2 and its perceival by the resistance protein I-2.
Dr Fleur Gawehns, PhD student - CBSG2012: Functional characterisation of Fusarium secreted effector proteins in disease and resistance
Dr ir. Ewa Lukasik, PhD student -Bioexploit: "Role of the ATPase activity of the NBS domain of R proteins and the identification of interacting proteins.
drs Vladimir Krasikov, Postdoc -UvA: Identifying the role of the Xsp10 protein for resistance and disease to Fusarium oxysporum.
dr. ir. Harrold van den Burg, Postdoc, NWO Veni: Eludidating the role of SUMO modification in plant defence signalling.
ing Marianne de Vroomen Technician, involved in most ongoing projects.
Drs Mobien Kasiem PhD student: PhD student: Structure and function of tomato disease resistance proteins.
Dr Gerben van Ooijen PhD student -NWO open program: Structure and function of tomato disease resistance proteins.
Dr Klaas Jan de Vries, Postdoc - CBSG 2008: Targeted proteomics of signalosomes mediating pathogen resistance.
Dr Wladimir Tameling PhD student: Disease resistance proteins of the NBS-LRR class, molecular switches of plant defence.
Dr Sergio de la Fuente van Bentem PhD student: Unfolding plant disease resistance, the involvement of HSP90 and its co-chaperone PP5 in I-2 mediated signalling.
Dr Jack Vossen, Postdoc: Characterisation of the I-2 signalosome.
Dr Sandra Elzinga, Postdoc: Characterisation of the Mi-1 signalosome.
-
Seminars and Patents (also publications <2002)
Invited speaker
Selection from the last ten years
- Identification of host targets of Fusarium effector proteins provides insight in disease resistance and susceptibility. 17 June 2022, IBMCP symposium at CSIC-Universitat Politècnica de València, Spain.
- Molecular aspects of endophyte-mediated resistance induced by Fusarium oxysporum. 15 December, 2021 (on line). Institute Colloquium, ETH, Zurich, Switzerland
- Molecular aspects of endophyte-mediated resistance induced by Fusarium oxysporum. 26 November 2021 (on line). Colloquium, Phytopathology, WUR, Wageningen, NL
- Molecular aspects of endophyte-mediated resistance induced by Fusarium oxysporum. VISCEA 4thInternational Conference “Plant Biotic Stresses & Resistance Mechanisms IV” February 19-20, 2020 Technische Universität Wien, Wien, Austria
- Combatting vascular diseases: identifying the guardians of the xylem. December 9 2019 Invited lecture CRAG, Barcelona, Spain
- Good fungi turning bad – what is the difference between beneficial and pathogenic Fusarium oxysporum strains? 25-28 September 2019, New Zealand Microbial Society Conference. Palmerston North, New Zealand.
- The Fusarium oxysporum effector Six8 manipulates plant immunity via the transcriptional co-repressors TPL and TPR1. 25 September 2019, AgResearch, Palmerston North, New Zealand.
- The good, the bad and the ugly: Unraveling the molecular basis of endophyte mediated resistance in tomato triggered by Fusarium oxysporum. September 20 2019 Invited lecture Copenhagen University, Copenhagen, Denmark
- The good, the bad and the ugly: Unraveling the molecular basis of endophyte mediated resistance in tomato triggered by Fusarium oxysporum. June 6 2019, Symposium Molecular Plant Pathology; Past and Future, Amsterdam, The Netherlands
- The CC domains of NLR-type pathogen receptors play essential roles in oligomerization, network formation and immune signalling. July 14-18 2019, MPMI XVIII, Glasgow, Scotland
- The good, the bad and the ugly : Unraveling the molecular basis of endophyte mediated resistance in tomato triggered by Fusarium oxysporum. March 21 2019, Top Lectures IBL-Leiden, the Netherlands
- The good, the bad and the ugly; unravelling the molecular basis of beneficial, pathogenic and commensal interactions between Fusarium oxysporum and tomato. October 26, 2018. Oxford University, Department of Plant Sciences, UK
- The good, the bad and the ugly; unravelling the molecular basis of beneficial, pathogenic and commensal interactions between Fusarium oxysporum and tomato. October 10th, 2018. "The XXIVth Minisymposium on Plant Biology", Koln, Germany
- The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity facilitating cell-to-cell movement of Avr2. July 2-3, 2018, 3rd International Conference “Plant Biotic Stresses & Resistance Mechanisms” Vienna Austria
- The good, the bad and the ugly; unravelling the molecular basis of beneficial, pathogenic and commensal interactions between Fusarium oxysporum and tomato. June 14, 2018 , KEYS symposium.“Harnessing plant-microbe symbiosis to revolutionize agriculture”, Wageningen, NL
- A tale of two effector proteins -How a pair of fusarium effector proteins manipulates tomato. January 20 2018, Hamburg University. Hamburg, Germany
- The Fusarium oxysporum Avr2-Six5 effector pair alters the exclusion selectivity of plasmodesmata. December 1, 2017. Massey University, Palmerston North New Zealand.
- The good, the bad and the ugly - genetic requirements of beneficial, pathogenic and commensal Fusarium oxysporum strains for colonization of tomato plants. 2-23 Nov 2017. NZSM annual conference 2017: our well-being & our microbes. Auckland, New Zealand
- The Fusarium oxysporum effector Six8 manipulates plant immunity through association with the transcriptional co-repressors TPL and TPR1. 28 August 2017, Shanghai Center for Plant Stress Biology, Shanghai, P.R. China.
- The Fusarium oxysporum Avr2-Six5 effector pair alters the exclusion selectivity of plasmodesmata and targets PTI signaling. 23 August 2017. Nanjing Agricultural University, Lab of Molecular Plant Virology, Nanjing. P.R. China.
- The Fusarium oxysporum Avr2-Six5 effector pair alters exclusion selectivity of plasmodesmata, 12 July 2017, EMBO Workshop; Intracellular Communication in development and disease, Harnack House, Berlin, Germany
- Functional analysis of the Fusarium Avr2 effector protein. 24 March 2017, Gregor Mendel Insitute Vienna Austria.
- The role of Fusarium effectors in virulence and NLR-mediated innate immunity. December 1 2016, Institut für Biologie/Genetik Martin-Luther-Universität, Halle (Saale), Germany
- Functional analysis of the Fusarium Avr2 effector protein. October 26 016, Sainsbury Laboratory, Norwich United Kingdom
- Gene-for-gene resistance - old dogma’s new insights: NLR’s, sentinels of plant immunity. TSL Summer School: Plant Microbe Interactions. 17-28 August 2015. TSL Norwich, United Kingdom
- The Fusarium oxysporum effector Six8 manipulates plant immunity through association with the transcriptional co-repressors TPL and TPR1. Annual Conference of the SUSTAIN COST Action (FA1208) 17-19 February 2016 Banyuls, France
- The role of Fusarium effectors in virulence and NLR-mediated innate immunity. New Phytologist workshop; “The apoplast as battleground for plant-microbe interactions”. 9 th– 10 st July 2015, Castle Rauischholzhausen, Gießen, Germany
- The Fusarium oxysporum effector Six8 manipulates plant immunity through association with the transcriptional co-repressors TPL and TPR1. International Conference on “Plant Biotic Stresses & Resistance Mechanisms II” July 2 - 4, 2015 Vienna, Austria
- The NLR immune receptors I-2 and Rx1 are DNA deforming proteins. 13-14 April 2015 De Wereld, Lunteren, The Netherlands
- The NLR immune receptors I-2 and Rx1 are DNA deforming proteins. MPI symposium ‘NLR Biology in Plants and Animals’, May 3-6, 2015 Schloss Ringberg Munich, Germany
- The NLR immune receptors I-2 and Rx1 are DNA deforming proteins. 15 Jan 2015 University of Copenhagen Copenhagen, Denmark
- The Fusarium oxysporum effector Six8 manipulates plant immunity through association with the transcriptional co-repressors TPL and TPR1. MINI-SYMPOSIUM: Wageningen University 24 Nov 2014, Wageningen Campus - Netherlands
- The tomato-fusarium pathosystem: a game of hide and seek, 16th IS-MPMI July 2014 Rhodos Greece
- The Fusarium oxysporum effector Six8 manipulates plant immunity through association with the transcriptional co-repressors TPL and TPR1. COST WORKSHOP: Structure-guided investigation of effector function, action and recognition. 10 – 12 Sept 2014, Bucharest, Romania
- De wapenwedloop tussen planten en hun belagers, een strijd om leven en dood. 5 March 2014 Spui lezing Amsterdam
- The role of Fusarium effectors in NLR-mediated innate immunity. XV International Congress on Molecular Plant-Microbe Interactions (IS-MPMI) July 29-August 2, 2012, Kyoto, Japan
- How to build a pathogen detector; structural basis of NB-LRR function. EMBO practical course 18-29 june 2012 TSL, Norwich, UK
- The role of Fusarium effectors in NLR-mediated plant innate immunity. 9 April 2012, Plant Biology Department, University of Fribourg, Fribourg Switzerland.
- The role of pathogen effectors in NLR-mediated plant innate immunity. International Meeting of the Collaborative Research Center SFB 648 "Communication in Plants and their Responsesto the Environment" May 19-22, 2011, Halle (Saale), Germany
- Molecular co-evolution in the tomato-Fusarium pathosystem. October 8, 2010, Justus Liebig University , Giessen, Germany
- Functional analysis of Fusarium oxysporum f.sp. lycopersici effector proteins. DFG funded colloquium: 'Microbial reprogramming of plant cell development' priority project SPP1212 'Plant-Micro' June 2010, Freising, Germany
- The arms race between effectors of Fusarium oxysporum and resistance proteins of tomato. Seminar series School of Biological and Biomedical Sciences, May 2010. Durham University, Durham, UK
- How to resist a resistance protein? British Society for Plant Pathology, Presidential Meeting, 2009, Oxford, UK
- How to resist tomato resistance proteins? XIV International Congress on Molecular Plant-Microbe Interactions. July 19-23, 2009, Quebec, Canada.
- Resistance proteins: scouts of the plant innateimmune system. 18th EUCARPIA congress, "Modern Variety Breeding for Present and Future Needs" 9-12 September 2008, Valencia, Spain
- Structure-function analysis of plant NB-LRR disease resistance proteins. SFB 670 " Structure Function and Evolution of Innate immunity" 22-23 September 2008, Max-Planck-Institut für Züchtungsforschung, Koln, Germany
- Resistance proteins: scouts of the plant innate immune system. EPS/Bioexploit Summerschool "On the evolution of plant-pathogen interactions: from principles to practices" 18-20 June 2008, Wageningen, the Netherlands.
- Resistance proteins: scouts of the plant innate immune system. 4th EPSO Conference, 22-26 June 2008 , Presqu'ile de Giens (Côte d'Azur), France
- Molecular aspects of I-2 mediated resistanceto Fusarium oxysporum . Seminar series. May 2008, Max-Planck-Institut für Züchtungsforschung, Koln, Germany
- Function of the Nucleotide Binding Domain for R Proteins. Keystone Symposium on Plant Innate Immunity, Feb 2008 Keystone Resort, Keystone, Colorado, USA
Publications arising from my PhD and Postdoc period, for recent papers see Tab "publications"
Papers <2002
- C. F. de Jong, F. L. W. Takken , X. Cai, P. J. G. M. de Wit and M. H. A.J. Joosten (2002)Attenuation of Cf -mediated defenseresponses atelevated temperatures correlates withadecrease in elicitor-binding sites.Molecular Plant-Microbe Interactions 15: 10. 1040-9.
- P. J. G. M. de Wit, B. F. Brandwagt, H. A. Van den Burg, X. Cai, R. A. L. Van der Hoorn, C.F. De Jong, J. van 't Klooster, M. J. D. de Kock, M. Kruijt, W. H. Lindhout, R. Luderer, F. L. W. Takken , N. Westerink, J.J. M. vervoort and M. H. A. J. Joosten (2002) The molecular basis of co-evolution between Cladosporium fulvum and tomato. Antonie vanLeeuwenhoek 81: 409-412
- R. Luderer, F. L. W. Takken , P.J. G.m.Witand M. H. A. J. Joosten (2002) Cladosporiumfulvum overcomes Cf-2 -mediated resistance by producing truncated AVR2 elicitor proteins. Molecular Microbiology 45: 3. 875-84.
- W. I. L. Tameling, S.D. Elzinga,P.S. Darmin, J. H. Vossen, F. L. W. Takken , M.A.Haring and B. J. C. Cornelissen (2002) The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins withATPase activity. Plant Cell 14: 11. 2929-39.
- X. Cai, F. L. W. Takken , M. H. A. J. Joosten and P.J. G. M. De Wit(2001) Specific recognition of Avr4 and Avr9 result in distinct patternsof hypersensitive cell death in tomato, but similar patterns ofdefence-related geneepxression. Molecular Plant Pathology 2: 2. 77-86
- B. F. Brandwagt, L. A.Mesbah, F. L. W. Takken , P. L. Laurent, T. J. A. Kneppers, J. Hille and H. J. J. Nijkamp (2000) A longevity assurance gene homolog of tomatomediates resistanceto Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc Natl Acad Sci USA 97:9.4961-6.
- F.L. W.Takken and M. H. A. J.Joosten (2000) Plant resistance genes: their structure, function and evolution. European Journal of Plant Pathology 106: 8. 699-713
- F. L. W. Takken, R. Luderer, S. H. E. J. Gabriëls, N.Westerink, R. Lu, P.J.G. M. de Wit and M.H.A.J.Joosten (2000) A functional cloning strategy, based on a binaryPVX-expression vector, toisolate HR-inducing cDNAsof plant pathogens. The Plant Journal 24: 2). 275-283
- L. A.Mesbah, T.J. A. Kneppers, F. L. W. Takken, P. Laurent,J.Hille and H. J. J. Nijkamp (1999) Geneticand physical analysis of aYAC contig spanning the fungal diseaseresistancelocus Asc of tomato ( Lycopersicon esculentum ). Molecular and General Genetics 261 (1): 50-57
- F.L. W. Takken , M. Thomas, M.H. A. J. Joosten, C. Golstein, N.Westerink, J. Hille, H. J. J. Nijkamp, P. J. G. M. DeWit and J. D. G. Jones (1999) A second gene at the tomato Cf-4 locus confers resistance to Cladosporium fulvum through recognition of anovel avirulence determinant. The Plant Journal 20: 3. 279-288
- F.L. W. Takken , D. Schipper, H. J. J. Nijkamp and J.Hille (1998)Identification and Ds-tagged isolation of a new gene at the Cf-4 locus of tomato involved in disease resistance to Cladosporium fulvum race 5. The Plant Journal 14: 401-411
patent applications
- Takken, F.L.W., &Wit, P.J.G.M.de (10-01-2002). Elicitor from Cladosporium . no WO02/02787.
- Turk, S.C.H.J., Takken, F.L.W., Jong, C.F. de, Joosten, M.H.A.J.,& Wit, P.J.G.M. de (02-01-2003). Nucleotide sequences involved in plantdisease resistance.no01202420.4./ WO03000930
- Joosten, M.H.A.J., Jong, C.F. de, Wit, P.J.G.M. de, Takken, F.L.W., & Turk, S.C.H.J. (17-03-2004). Nucleotide sequences involved in plantresistance. no EP1397515
-
Biography
Biography
Frank Takken (1969) is interested in how plants defend themselves against pathogens. Plants are able to sense the presence of invading microbes. Recognition subsequently results in activation of the plant innate immune system. In this process, resistance proteins play a key role as they mediate pathogen recognition and trigger the activation of downstream signaling cascades that halt the pathogen and thereby prevent disease. An example of a succesfull immune response is shown in the figure below where the tomato Immunity-2 ( I-2) gene halts ingress of the pathogenic fungus Fusarium oxysporum.
Resistance genes come in different flavors, but the majority encodes intracellular tri-partite proteins that contain a central nucleotide-binding domain. One of Frank's major research interests is the role the distinct domains in these proteins have for their function. Structure-function analysis in combination with 3D modeling of the different subdomain is used to predict mutations that will have specific effects on the activity of these proteins. These mutants are subsequently analyzed for altered biochemical properties, differences in inter- and intramolecular interactions as well as for their effect on disease resistance.
To examine how resistance proteins activate downstream signaling it is essential to know their interacting partners. Using various screens, candidate interacting proteins, and recently also DNA, have been identified and these interactors are analyzed for their involvement in plant defense. Part of this research is funded by an NWO VICI grant that was awarded in 2015. Besides resistance genes, I also have a long standing interest to resolve the molecular basis of host susceptibility and the induced resistance responses triggered by biocontrol fungi.
Frank is appointed as Associate professor in the Molecular Plant Pathology group at the Swammerdam Institute for Life Sciences and was from 2012-2019 Scientific Advisor for Scienza Biotechnology. Previously he worked as postdoc at the Phytopathology group at Wageningen University. During this time he worked on a self-written project at the biotech company Keygene NV in Wageningen. In that project he focused on the identification of genes that are transcriptionally regulated during the onset of plant defense signaling. He received his PhD in 1999 from the Free University of Amsterdam for his pioneering work on the isolation of resistance genes from tomato.
A resistant tomato plant carrying I-2 (left) and susceptible one (right side) infected with Fusarium oxysporum f.sp. lycopersici. -
Publications
2024
 Aalders, T. R., de Sain, M., Gawehns, F., Oudejans, N., Jak, Y. D., Dekker, H. L., Rep, M., van den Burg, H. A., & Takken, F. L. W. (2024). Specific members of the TOPLESS family are susceptibility genes for Fusarium wilt in tomato and Arabidopsis. Plant Biotechnology Journal, 22(1), 248-261. https://doi.org/10.1111/pbi.14183 [details]
Aalders, T. R., de Sain, M., Gawehns, F., Oudejans, N., Jak, Y. D., Dekker, H. L., Rep, M., van den Burg, H. A., & Takken, F. L. W. (2024). Specific members of the TOPLESS family are susceptibility genes for Fusarium wilt in tomato and Arabidopsis. Plant Biotechnology Journal, 22(1), 248-261. https://doi.org/10.1111/pbi.14183 [details]
2023
 Blekemolen, M. C., Liu, Z., Stegman, M., Zipfel, C., Shan, L., & Takken, F. L. W. (2023). The PTI-suppressing Avr2 effector from Fusarium oxysporum suppresses mono-ubiquitination and plasma membrane dissociation of BIK1. Molecular Plant Pathology, 24(10), 1273-1286. https://doi.org/10.1111/mpp.13369 [details]
Blekemolen, M. C., Liu, Z., Stegman, M., Zipfel, C., Shan, L., & Takken, F. L. W. (2023). The PTI-suppressing Avr2 effector from Fusarium oxysporum suppresses mono-ubiquitination and plasma membrane dissociation of BIK1. Molecular Plant Pathology, 24(10), 1273-1286. https://doi.org/10.1111/mpp.13369 [details]- Talbi, N., Blekemolen, M. C., Janevska, S., Zendler, D. P., Van Tilbeurgh, H., Fudal, I., & Takken, F. L. W. (2023). Facilitation of symplastic effector protein mobility by paired effectors is conserved in different classes of fungal pathogens. Molecular Plant-Microbe Interactions. Advance online publication. https://doi.org/10.1094/MPMI-07-23-0103-FI
- Vermeulen, A., Takken, F. L. W., & Sánchez-Camargo, V. A. (2023). Translation Arrest: A Key Player in Plant Antiviral Response. Genes, 14(6), Article 1293. https://doi.org/10.3390/genes14061293
- Zhang, W., Planas-Marquès, M., Mazier, M., Šimkovicová, M., Rocafort, M., Mantz, M., Huesgen, P. F., Takken, F. L. W., Stintzi, A., Schaller, A., Coll, N. S., & Valls, M. (in press). The tomato P69 subtilase family is involved in resistance to bacterial wilt. Plant Journal. https://doi.org/10.1111/tpj.16613
2022
 Blekemolen, M. C., Cao, L., Tintor, N., de Groot, T., Papp, D., Faulkner, C., & Takken, F. L. W. (2022). The primary function of Six5 of Fusarium oxysporum is to facilitate Avr2 activity by together manipulating the size exclusion limit of plasmodesmata. Frontiers in Plant Science, 13, Article 910594. https://doi.org/10.3389/fpls.2022.910594 [details]
Blekemolen, M. C., Cao, L., Tintor, N., de Groot, T., Papp, D., Faulkner, C., & Takken, F. L. W. (2022). The primary function of Six5 of Fusarium oxysporum is to facilitate Avr2 activity by together manipulating the size exclusion limit of plasmodesmata. Frontiers in Plant Science, 13, Article 910594. https://doi.org/10.3389/fpls.2022.910594 [details] Tintor, N., Nieuweboer, G. A. M., Bakker, I. A. W., & Takken, F. L. W. (2022). The Intracellularly Acting Effector Foa3 Suppresses Defense Responses When Infiltrated Into the Apoplast. Frontiers in Plant Science, 13, Article 813181. https://doi.org/10.3389/fpls.2022.813181 [details]
Tintor, N., Nieuweboer, G. A. M., Bakker, I. A. W., & Takken, F. L. W. (2022). The Intracellularly Acting Effector Foa3 Suppresses Defense Responses When Infiltrated Into the Apoplast. Frontiers in Plant Science, 13, Article 813181. https://doi.org/10.3389/fpls.2022.813181 [details]
2021
 Coleman, A. D., Maroschek, J., Raasch, L., Takken, F. L. W., Ranf, S., & Hückelhoven, R. (2021). The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytologist, 229(6), 3453-3466. Advance online publication. https://doi.org/10.1111/nph.17122 [details]
Coleman, A. D., Maroschek, J., Raasch, L., Takken, F. L. W., Ranf, S., & Hückelhoven, R. (2021). The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytologist, 229(6), 3453-3466. Advance online publication. https://doi.org/10.1111/nph.17122 [details] Constantin, M. E., Fokkens, L., de Sain, M., Takken, F. L. W., & Rep, M. (2021). Number of Candidate Effector Genes in Accessory Genomes Differentiates Pathogenic From Endophytic Fusarium oxysporum Strains. Frontiers in Plant Science, 12, Article 761740. https://doi.org/10.3389/fpls.2021.761740 [details]
Constantin, M. E., Fokkens, L., de Sain, M., Takken, F. L. W., & Rep, M. (2021). Number of Candidate Effector Genes in Accessory Genomes Differentiates Pathogenic From Endophytic Fusarium oxysporum Strains. Frontiers in Plant Science, 12, Article 761740. https://doi.org/10.3389/fpls.2021.761740 [details] Richard, M. M. S., Knip, M., Schachtschabel, J., Beijaert, M. S., & Takken, F. L. W. (2021). Perturbation of nuclear–cytosolic shuttling of Rx1 compromises extreme resistance and translational arrest of potato virus X transcripts. Plant Journal, 106(2), 468-479. https://doi.org/10.1111/tpj.15179 [details]
Richard, M. M. S., Knip, M., Schachtschabel, J., Beijaert, M. S., & Takken, F. L. W. (2021). Perturbation of nuclear–cytosolic shuttling of Rx1 compromises extreme resistance and translational arrest of potato virus X transcripts. Plant Journal, 106(2), 468-479. https://doi.org/10.1111/tpj.15179 [details] de Lamo, F. J., Spijkers, S. B., & Takken, F. L. W. (2021). Protection to tomato wilt disease conferred by the non-pathogen Fusarium oxysporum Fo47 is more effective than that conferred by avirulent strains. Phytopathology, 111(2), 253-257. Advance online publication. https://doi.org/10.1094/PHYTO-04-20-0133-R [details]
de Lamo, F. J., Spijkers, S. B., & Takken, F. L. W. (2021). Protection to tomato wilt disease conferred by the non-pathogen Fusarium oxysporum Fo47 is more effective than that conferred by avirulent strains. Phytopathology, 111(2), 253-257. Advance online publication. https://doi.org/10.1094/PHYTO-04-20-0133-R [details] de Lamo, F. J., Šimkovicová, M., Fresno, D. H., de Groot, T., Tintor, N., Rep, M., & Takken, F. L. W. (2021). Pattern-triggered immunity restricts host colonization by endophytic fusaria, but does not affect endophyte-mediated resistance. Molecular Plant Pathology, 22(2), 204-215. https://doi.org/10.1111/mpp.13018 [details]
de Lamo, F. J., Šimkovicová, M., Fresno, D. H., de Groot, T., Tintor, N., Rep, M., & Takken, F. L. W. (2021). Pattern-triggered immunity restricts host colonization by endophytic fusaria, but does not affect endophyte-mediated resistance. Molecular Plant Pathology, 22(2), 204-215. https://doi.org/10.1111/mpp.13018 [details]
2020
 Constantin, M. E., Vlieger, B. V., Takken, F. L. W., & Rep, M. (2020). Diminished Pathogen and Enhanced Endophyte Colonization upon CoInoculation of Endophytic and Pathogenic Fusarium Strains. Microorganisms, 8(4), Article 544. https://doi.org/10.3390/microorganisms8040544 [details]
Constantin, M. E., Vlieger, B. V., Takken, F. L. W., & Rep, M. (2020). Diminished Pathogen and Enhanced Endophyte Colonization upon CoInoculation of Endophytic and Pathogenic Fusarium Strains. Microorganisms, 8(4), Article 544. https://doi.org/10.3390/microorganisms8040544 [details] Constantin, M. E., de Lamo, F. J., Rep, M., & Takken, F. L. W. (2020). From laboratory to field: applying the Fo47 biocontrol strain in potato fields. European Journal of Plant Pathology, 158(3), 645–654. https://doi.org/10.1007/s10658-020-02106-6 [details]
Constantin, M. E., de Lamo, F. J., Rep, M., & Takken, F. L. W. (2020). From laboratory to field: applying the Fo47 biocontrol strain in potato fields. European Journal of Plant Pathology, 158(3), 645–654. https://doi.org/10.1007/s10658-020-02106-6 [details] Richard, M. M. S., Knip, M., Aalders, T., Beijaert, M. S., & Takken, F. L. W. (2020). Unlike Many Disease Resistances, Rx1-Mediated Immunity to Potato Virus X Is Not Compromised at Elevated Temperatures. Frontiers in Genetics, 11, Article 417. https://doi.org/10.3389/fgene.2020.00417 [details]
Richard, M. M. S., Knip, M., Aalders, T., Beijaert, M. S., & Takken, F. L. W. (2020). Unlike Many Disease Resistances, Rx1-Mediated Immunity to Potato Virus X Is Not Compromised at Elevated Temperatures. Frontiers in Genetics, 11, Article 417. https://doi.org/10.3389/fgene.2020.00417 [details] Sukarta, O. C. A., Townsend, P. D., Llewelyn, A., Dixon, C. H., Slootweg, E. J., Pålsson, L-O., Takken, F. L. W., Goverse, A., & Cann, M. J. (2020). A DNA-Binding Bromodomain-Containing Protein Interacts with and Reduces Rx1-Mediated Immune Response to Potato Virus X. Plant communications, 1(4), 100086. https://doi.org/10.1016/j.xplc.2020.100086 [details]
Sukarta, O. C. A., Townsend, P. D., Llewelyn, A., Dixon, C. H., Slootweg, E. J., Pålsson, L-O., Takken, F. L. W., Goverse, A., & Cann, M. J. (2020). A DNA-Binding Bromodomain-Containing Protein Interacts with and Reduces Rx1-Mediated Immune Response to Potato Virus X. Plant communications, 1(4), 100086. https://doi.org/10.1016/j.xplc.2020.100086 [details] Tintor, N., Paauw, M., Rep, M., & Takken, F. L. W. (2020). The root invading pathogen Fusarium oxysporum targets pattern-triggered immunity using both cytoplasmic and apoplastic effectors. New Phytologist, 227(5), 1479-1492. https://doi.org/10.1111/nph.16618 [details]
Tintor, N., Paauw, M., Rep, M., & Takken, F. L. W. (2020). The root invading pathogen Fusarium oxysporum targets pattern-triggered immunity using both cytoplasmic and apoplastic effectors. New Phytologist, 227(5), 1479-1492. https://doi.org/10.1111/nph.16618 [details] Xue, J.-Y., Takken, F. L. W., Nepal, M. P., Maekawa, T., & Shao, Z.-Q. (2020). Editorial: Evolution and Functional Mechanisms of Plant Disease Resistance. Frontiers in Genetics, 11, Article 593240. https://doi.org/10.3389/fgene.2020.593240 [details]
Xue, J.-Y., Takken, F. L. W., Nepal, M. P., Maekawa, T., & Shao, Z.-Q. (2020). Editorial: Evolution and Functional Mechanisms of Plant Disease Resistance. Frontiers in Genetics, 11, Article 593240. https://doi.org/10.3389/fgene.2020.593240 [details] de Lamo, F. J., & Takken, F. L. W. (2020). Biocontrol by Fusarium oxysporum Using Endophyte-Mediated Resistance. Frontiers in Plant Science, 11, Article 37. https://doi.org/10.3389/fpls.2020.00037 [details]
de Lamo, F. J., & Takken, F. L. W. (2020). Biocontrol by Fusarium oxysporum Using Endophyte-Mediated Resistance. Frontiers in Plant Science, 11, Article 37. https://doi.org/10.3389/fpls.2020.00037 [details]
2019
 Constantin, M. E., de Lamo, F. J., Vlieger, B. V., Rep, M., & Takken, F. L. W. (2019). Endophyte-Mediated Resistance in Tomato to Fusarium oxysporum Is Independent of ET, JA, and SA. Frontiers in Plant Science, 10, Article 979. https://doi.org/10.3389/fpls.2019.00979 [details]
Constantin, M. E., de Lamo, F. J., Vlieger, B. V., Rep, M., & Takken, F. L. W. (2019). Endophyte-Mediated Resistance in Tomato to Fusarium oxysporum Is Independent of ET, JA, and SA. Frontiers in Plant Science, 10, Article 979. https://doi.org/10.3389/fpls.2019.00979 [details] Knip, M., Richard, M. M. S., Oskam, L., van Engelen, H. T. D., Aalders, T., & Takken, F. L. W. (2019). Activation of immune receptor Rx1 triggers distinct immune responses culminating in cell death after 4 hours. Molecular Plant Pathology, 20(4), 575-588. Advance online publication. https://doi.org/10.1111/mpp.12776 [details]
Knip, M., Richard, M. M. S., Oskam, L., van Engelen, H. T. D., Aalders, T., & Takken, F. L. W. (2019). Activation of immune receptor Rx1 triggers distinct immune responses culminating in cell death after 4 hours. Molecular Plant Pathology, 20(4), 575-588. Advance online publication. https://doi.org/10.1111/mpp.12776 [details]- de Lamo, F. J., Constantin, M. E., Fresno, D. H., Boeren, S., Rep, M., & Takken, F. L. W. (2019). Corrigendum: Xylem Sap Proteomics Reveals Distinct Differences Between R Gene- and Endophyte-Mediated Resistance Against Fusarium Wilt Disease in Tomato. Frontiers in Microbiology, 10. https://doi.org/10.3389/fmicb.2019.01872
 van der Does, H. C., Constantin, M. E., Houterman, P. M., Takken, F. L. W., Cornelissen, B. J. C., Haring, M. A., van den Burg, H. A., & Rep, M. (2019). Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. European Journal of Plant Pathology, 154(1), 55-65. https://doi.org/10.1007/s10658-018-1596-3 [details]
van der Does, H. C., Constantin, M. E., Houterman, P. M., Takken, F. L. W., Cornelissen, B. J. C., Haring, M. A., van den Burg, H. A., & Rep, M. (2019). Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. European Journal of Plant Pathology, 154(1), 55-65. https://doi.org/10.1007/s10658-018-1596-3 [details]
2018
 Blekemolen, M. C., Tark-Dame, M., & Takken, F. L. W. (2018). Visualization and Quantification of Cell-to-cell Movement of Proteins in Nicotiana benthamiana. BIO-PROTOCOL, 8(24), Article e3114. https://doi.org/10.21769/BioProtoc.3114 [details]
Blekemolen, M. C., Tark-Dame, M., & Takken, F. L. W. (2018). Visualization and Quantification of Cell-to-cell Movement of Proteins in Nicotiana benthamiana. BIO-PROTOCOL, 8(24), Article e3114. https://doi.org/10.21769/BioProtoc.3114 [details] Cao, L., Blekemolen, M. C., Tintor, N., Cornelissen, B. J. C., & Takken, F. L. W. (2018). The Fusarium oxysporum Avr2-Six5 Effector Pair Alters Plasmodesmatal Exclusion Selectivity to Facilitate Cell-to-Cell Movement of Avr2. Molecular Plant, 11(5), 691-705. Advance online publication. https://doi.org/10.1016/j.molp.2018.02.011 [details]
Cao, L., Blekemolen, M. C., Tintor, N., Cornelissen, B. J. C., & Takken, F. L. W. (2018). The Fusarium oxysporum Avr2-Six5 Effector Pair Alters Plasmodesmatal Exclusion Selectivity to Facilitate Cell-to-Cell Movement of Avr2. Molecular Plant, 11(5), 691-705. Advance online publication. https://doi.org/10.1016/j.molp.2018.02.011 [details] Townsend, P. D., Dixon, C. H., Slootweg, E. J., Sukarta, O. C. A., Yang, A. W. H., Hughes, T. R., Sharples, G. J., Palsson, L-O., Takken, F. L. W., Goverse, A., & Cann, M. J. (2018). The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. The Journal of Biological Chemistry, 293(9), 3218-3233. https://doi.org/10.1074/jbc.RA117.000485 [details]
Townsend, P. D., Dixon, C. H., Slootweg, E. J., Sukarta, O. C. A., Yang, A. W. H., Hughes, T. R., Sharples, G. J., Palsson, L-O., Takken, F. L. W., Goverse, A., & Cann, M. J. (2018). The intracellular immune receptor Rx1 regulates the DNA-binding activity of a Golden2-like transcription factor. The Journal of Biological Chemistry, 293(9), 3218-3233. https://doi.org/10.1074/jbc.RA117.000485 [details] Wróblewski, T., Spiridon, L., Martin, E. C., Petrescu, A-J., Cavanaugh, K., Truco, M. J., Xu, H., Gozdowski, D., Pawłowski, K., Michelmore, R. W., & Takken, F. L. W. (2018). Genome-wide functional analyses of plant coiled-coil NLR-type pathogen receptors reveal essential roles of their N-terminal domain in oligomerization, networking, and immunity. PLoS Biology, 16(12), Article e2005821. https://doi.org/10.1371/journal.pbio.2005821 [details]
Wróblewski, T., Spiridon, L., Martin, E. C., Petrescu, A-J., Cavanaugh, K., Truco, M. J., Xu, H., Gozdowski, D., Pawłowski, K., Michelmore, R. W., & Takken, F. L. W. (2018). Genome-wide functional analyses of plant coiled-coil NLR-type pathogen receptors reveal essential roles of their N-terminal domain in oligomerization, networking, and immunity. PLoS Biology, 16(12), Article e2005821. https://doi.org/10.1371/journal.pbio.2005821 [details] de Lamo, F. J., Constantin, M. E., Fresno, D. H., Boeren, S., Rep, M., & Takken, F. L. W. (2018). Xylem Sap Proteomics Reveals Distinct Differences Between R Gene- and Endophyte-Mediated Resistance Against Fusarium Wilt Disease in Tomato. Frontiers in Microbiology, 9, Article 2977. https://doi.org/10.3389/fmicb.2018.02977 [details]
de Lamo, F. J., Constantin, M. E., Fresno, D. H., Boeren, S., Rep, M., & Takken, F. L. W. (2018). Xylem Sap Proteomics Reveals Distinct Differences Between R Gene- and Endophyte-Mediated Resistance Against Fusarium Wilt Disease in Tomato. Frontiers in Microbiology, 9, Article 2977. https://doi.org/10.3389/fmicb.2018.02977 [details]
2017
 Acevedo-Garcia, J., Gruner, K., Reinstädler, A., Kemen, A., Kemen, E., Cao, L., Takken, F. L. W., Reitz, M. U., Schäfer, P., O'Connell, R. J., Kusch, S., Kuhn, H., & Panstruga, R. (2017). The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Scientific Reports, 7, Article 9319. https://doi.org/10.1038/s41598-017-07188-7 [details]
Acevedo-Garcia, J., Gruner, K., Reinstädler, A., Kemen, A., Kemen, E., Cao, L., Takken, F. L. W., Reitz, M. U., Schäfer, P., O'Connell, R. J., Kusch, S., Kuhn, H., & Panstruga, R. (2017). The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Scientific Reports, 7, Article 9319. https://doi.org/10.1038/s41598-017-07188-7 [details] Di, X., Cao, L., Hughes, R. K., Tintor, N., Banfield, M. J., & Takken, F. L. W. (2017). Structure-function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytologist, 216(3), 897-914. Advance online publication. https://doi.org/10.1111/nph.14733 [details]
Di, X., Cao, L., Hughes, R. K., Tintor, N., Banfield, M. J., & Takken, F. L. W. (2017). Structure-function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytologist, 216(3), 897-914. Advance online publication. https://doi.org/10.1111/nph.14733 [details] Di, X., Gomila, J., & Takken, F. L. W. (2017). Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the susceptibility of tomato to Fusarium oxysporum. Molecular Plant Pathology, 18(7), 1024–1035 . https://doi.org/10.1111/mpp.12559 [details]
Di, X., Gomila, J., & Takken, F. L. W. (2017). Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the susceptibility of tomato to Fusarium oxysporum. Molecular Plant Pathology, 18(7), 1024–1035 . https://doi.org/10.1111/mpp.12559 [details] Richard, M. M. S., & Takken, F. L. W. (2017). Plant Autoimmunity: When Good Things Go Bad. Current Biology, 27(9), R361-R363. https://doi.org/10.1016/j.cub.2017.03.039 [details]
Richard, M. M. S., & Takken, F. L. W. (2017). Plant Autoimmunity: When Good Things Go Bad. Current Biology, 27(9), R361-R363. https://doi.org/10.1016/j.cub.2017.03.039 [details]
2016
 Di, X., Gomila, J., Ma, L., van den Burg, H. A., & Takken, F. L. W. (2016). Uptake of the Fusarium Effector Avr2 by Tomato Is Not a Cell Autonomous Event. Frontiers in Plant Science, 7, Article 1915. https://doi.org/10.3389/fpls.2016.01915 [details]
Di, X., Gomila, J., Ma, L., van den Burg, H. A., & Takken, F. L. W. (2016). Uptake of the Fusarium Effector Avr2 by Tomato Is Not a Cell Autonomous Event. Frontiers in Plant Science, 7, Article 1915. https://doi.org/10.3389/fpls.2016.01915 [details] Di, X., Takken, F. L. W., & Tintor, N. (2016). How Phytohormones Shape Interactions between Plants and the Soil-Borne Fungus Fusarium oxysporum. Frontiers in Plant Science, 7, Article 170. https://doi.org/10.3389/fpls.2016.00170 [details]
Di, X., Takken, F. L. W., & Tintor, N. (2016). How Phytohormones Shape Interactions between Plants and the Soil-Borne Fungus Fusarium oxysporum. Frontiers in Plant Science, 7, Article 170. https://doi.org/10.3389/fpls.2016.00170 [details] Fenyk, S., Dixon, C. H., Gittens, W. H., Townsend, P. D., Sharples, G. J., Pålsson, L. O., Takken, F. L. W., & Cann, M. J. (2016). The Tomato Nucleotide-binding Leucine-rich Repeat (NLR) Immune Receptor I-2 Couples DNA-Binding to Nucleotide-Binding Domain Nucleotide Exchange. The Journal of Biological Chemistry, 291(3), 1137-1147. Advance online publication. https://doi.org/10.1074/jbc.M115.698589 [details]
Fenyk, S., Dixon, C. H., Gittens, W. H., Townsend, P. D., Sharples, G. J., Pålsson, L. O., Takken, F. L. W., & Cann, M. J. (2016). The Tomato Nucleotide-binding Leucine-rich Repeat (NLR) Immune Receptor I-2 Couples DNA-Binding to Nucleotide-Binding Domain Nucleotide Exchange. The Journal of Biological Chemistry, 291(3), 1137-1147. Advance online publication. https://doi.org/10.1074/jbc.M115.698589 [details] Peng, H-C., Mantelin, S., Hicks, G. R., Takken, F. L. W., & Kaloshian, I. (2016). The Conformation of the Plasma Membrane-Localized SlSERK1 - Mi-1.2 Complex is Altered by a Potato Aphid Derived Effector. Plant Physiology, 171(3), 2211-2222. Advance online publication. https://doi.org/10.1104/pp.16.00295 [details]
Peng, H-C., Mantelin, S., Hicks, G. R., Takken, F. L. W., & Kaloshian, I. (2016). The Conformation of the Plasma Membrane-Localized SlSERK1 - Mi-1.2 Complex is Altered by a Potato Aphid Derived Effector. Plant Physiology, 171(3), 2211-2222. Advance online publication. https://doi.org/10.1104/pp.16.00295 [details]
2015
 Fenyk, S., Townsend, P. D., Dixon, C. H., Spies, G. B., de San Eustaquio Campillo, A., Slootweg, E. J., Westerhof, L. B., Gawehns, F. K. K., Knight, M. R., Sharples, G. J., Goverse, A., Pålsson, L. O., Takken, F. L. W., & Cann, M. J. (2015). The Potato Nucleotide-binding Leucine-rich Repeat (NLR) Immune Receptor Rx1 Is a Pathogen-dependent DNA-deforming Protein. The Journal of Biological Chemistry, 290(41), 24945-24960. Advance online publication. https://doi.org/10.1074/jbc.M115.672121 [details]
Fenyk, S., Townsend, P. D., Dixon, C. H., Spies, G. B., de San Eustaquio Campillo, A., Slootweg, E. J., Westerhof, L. B., Gawehns, F. K. K., Knight, M. R., Sharples, G. J., Goverse, A., Pålsson, L. O., Takken, F. L. W., & Cann, M. J. (2015). The Potato Nucleotide-binding Leucine-rich Repeat (NLR) Immune Receptor Rx1 Is a Pathogen-dependent DNA-deforming Protein. The Journal of Biological Chemistry, 290(41), 24945-24960. Advance online publication. https://doi.org/10.1074/jbc.M115.672121 [details] Gawehns, F., Ma, L., Bruning, O., Houterman, P. M., Boeren, S., Cornelissen, B. J. C., Rep, M., & Takken, F. L. W. (2015). The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Frontiers in Plant Science, 6, Article 967. https://doi.org/10.3389/fpls.2015.00967 [details]
Gawehns, F., Ma, L., Bruning, O., Houterman, P. M., Boeren, S., Cornelissen, B. J. C., Rep, M., & Takken, F. L. W. (2015). The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Frontiers in Plant Science, 6, Article 967. https://doi.org/10.3389/fpls.2015.00967 [details] Ma, L., Houterman, P. M., Gawehns, F., Cao, L., Sillo, F., Richter, H., Clavijo-Ortiz, M. J., Schmidt, S. M., Boeren, S., Vervoort, J., Cornelissen, B. J. C., Rep, M., & Takken, F. L. W. (2015). The AVR2-SIX5 gene pair is required to activate I-2-mediated immunity in tomato. New Phytologist, 208(2), 507-518. Advance online publication. https://doi.org/10.1111/nph.13455 [details]
Ma, L., Houterman, P. M., Gawehns, F., Cao, L., Sillo, F., Richter, H., Clavijo-Ortiz, M. J., Schmidt, S. M., Boeren, S., Vervoort, J., Cornelissen, B. J. C., Rep, M., & Takken, F. L. W. (2015). The AVR2-SIX5 gene pair is required to activate I-2-mediated immunity in tomato. New Phytologist, 208(2), 507-518. Advance online publication. https://doi.org/10.1111/nph.13455 [details]
2014
- Gawehns, F., Houterman, P. M., Ait Ichou, F., Michielse, C. B., Hijdra, M., Cornelissen, B. J. C., Rep, M., & Takken, F. (2014). The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2 mediated cell death. Molecular Plant-Microbe Interactions, 27(4), 336-348. https://doi.org/10.1094/MPMI-11-13-0330-R [details]
- van Schie, C. C. N., & Takken, F. L. W. (2014). Susceptibility genes 101: how to be a good host. Annual Review of Phytopathology, 52, 551-581. https://doi.org/10.1146/annurev-phyto-102313-045854 [details]
2013
 Gawehns, F., Cornelissen, B. J. C., & Takken, F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microbial Biotechnology, 6(3), 223-229. Advance online publication. https://doi.org/10.1111/1751-7915.12023 [details]
Gawehns, F., Cornelissen, B. J. C., & Takken, F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microbial Biotechnology, 6(3), 223-229. Advance online publication. https://doi.org/10.1111/1751-7915.12023 [details] Ma, L., Cornelissen, B. J. C., & Takken, F. L. W. (2013). A nuclear localization for Avr2 from Fusarium oxysporum is required to activate the tomato resistance protein I-2. Frontiers in Plant Science, 4, 94. Advance online publication. https://doi.org/10.3389/fpls.2013.00094 [details]
Ma, L., Cornelissen, B. J. C., & Takken, F. L. W. (2013). A nuclear localization for Avr2 from Fusarium oxysporum is required to activate the tomato resistance protein I-2. Frontiers in Plant Science, 4, 94. Advance online publication. https://doi.org/10.3389/fpls.2013.00094 [details]- Ma, L., van den Burg, H. A., Cornelissen, B. J. C., & Takken, F. L. W. (2013). Molecular basis of effector recognition by plant NB-LRR proteins. In G. Sessa (Ed.), Molecular plant immunity (pp. 23-39). Wiley-Blackwell. https://doi.org/10.1002/9781118481431.ch2 [details]
 Pietraszewska-Bogiel, A., Lefebvre, B., Koini, M. A., Klaus-Heisen, D., Takken, F. L. W., Geurts, R., Cullimore, J. V., & Gadella, T. W. J. (2013). Interaction of Medicago truncatula Lysin Motif Receptor-Like Kinases, NFP and LYK3, Produced in Nicotiana benthamiana Induces Defence-Like Responses. PLoS ONE, 8(6), Article e65055. https://doi.org/10.1371/journal.pone.0065055 [details]
Pietraszewska-Bogiel, A., Lefebvre, B., Koini, M. A., Klaus-Heisen, D., Takken, F. L. W., Geurts, R., Cullimore, J. V., & Gadella, T. W. J. (2013). Interaction of Medicago truncatula Lysin Motif Receptor-Like Kinases, NFP and LYK3, Produced in Nicotiana benthamiana Induces Defence-Like Responses. PLoS ONE, 8(6), Article e65055. https://doi.org/10.1371/journal.pone.0065055 [details] Schmidt, S. M., Houterman, P. M., Schreiver, I., Ma, L., Amyotte, S., Chellappan, B., Boeren, S., Takken, F. L. W., & Rep, M. (2013). MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics, 14, 119. https://doi.org/10.1186/1471-2164-14-119 [details]
Schmidt, S. M., Houterman, P. M., Schreiver, I., Ma, L., Amyotte, S., Chellappan, B., Boeren, S., Takken, F. L. W., & Rep, M. (2013). MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics, 14, 119. https://doi.org/10.1186/1471-2164-14-119 [details]
2012
 Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., Xiao, W., Liu, Y., Chai, J., Takken, F. L. W., Schulze-Lefert, P., & Shen, Q. H. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathogens, 8(6), Article e10027. https://doi.org/10.1371/journal.ppat.1002752 [details]
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., Xiao, W., Liu, Y., Chai, J., Takken, F. L. W., Schulze-Lefert, P., & Shen, Q. H. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathogens, 8(6), Article e10027. https://doi.org/10.1371/journal.ppat.1002752 [details]- Lukasik-Shreepaathy, E., Slootweg, E., Richter, H., Goverse, A., Cornelissen, B. J. C., & Takken, F. L. W. (2012). Dual regulatory roles of the extended N terminus for activation of the tomato MI-1.2 resistance protein. Molecular Plant-Microbe Interactions, 25(8), 1045-1057. https://doi.org/10.1094/MPMI-11-11-0302 [details]
- Lukasik-Shreepaathy, E., Vossen, J. H., Tameling, W. I. L., de Vroomen, M. J., Cornelissen, B. J. C., & Takken, F. L. W. (2012). Protein-protein interactions as a proxy to monitor conformational changes and activation states of the tomato resistance protein I-2. Journal of Experimental Botany, 63(8), 3047-3060. https://doi.org/10.1093/jxb/ers021 [details]
- Ma, L., Lukasik, E., Gawehns, F., & Takken, F. L. W. (2012). The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. In M. D. Bolton, & B. P. H. J. Thomma (Eds.), Plant fungal pathogens: methods and protocols (pp. 61-74). (Methods in molecular biology; No. 835). Humana Press. https://doi.org/10.1007/978-1-61779-501-5_4 [details]
- Takken, F. L. W., & Goverse, A. (2012). How to build a pathogen detector: structural basis of NB-LRR function. Current Opinion in Plant Biology, 15(4), 375-384. https://doi.org/10.1016/j.pbi.2012.05.001 [details]
2011
- Krasikov, V., Dekker, H. L., Rep, M., & Takken, F. L. W. (2011). The tomato xylem sap protein XSP10 is required for full susceptibility to Fusarium wilt disease. Journal of Experimental Botany, 62(3), 963-973. https://doi.org/10.1093/jxb/erq327 [details]
- Maekawa, T., Cheng, W., Spiridon, L. N., Töller, A., Lukasik, E., Saijo, Y., Liu, P., Shen, Q. H., Micluta, M. A., Somssich, I. E., Takken, F. L. W., Petrescu, A. J., Chai, J., & Schulze-Lefert, P. (2011). Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. CELL Host & Microbe, 9(3), 187-199. https://doi.org/10.1016/j.chom.2011.02.008 [details]
- Mantelin, S., Peng, H. C., Li, B., Atamian, H. S., Takken, F. L. W., & Kaloshian, I. (2011). The receptor-like kinase SlSERK1 is required for Mi-1-mediated resistance to potato aphids in tomato. Plant Journal, 67(3), 459-471. https://doi.org/10.1111/j.1365-313X.2011.04609.x [details]
2010
- Ament, K., Krasikov, V., Allmann, S., Rep, M., Takken, F. L. W., & Schuurink, R. C. (2010). Methyl salicylate production in tomato affects biotic interactions. Plant Journal, 62(1), 124-134. https://doi.org/10.1111/j.1365-313X.2010.04132.x [details]
- Takken, F., & Rep, M. (2010). The arms race between tomato and Fusarium oxysporum. Molecular Plant Pathology, 11(2), 309-314. https://doi.org/10.1111/j.1364-3703.2009.00605.x [details]
- Takken, F., van Ooijen, G., Lukasik, E., Ma, L., Gawehns, F., Houterman, P., & Rep, M. (2010). How to resist a tomato resistance gene? In H. Antoun, T. Avis, L. Brisson, D. Prévost, & M. Trepanier (Eds.), Proceedings of the 14th International Congress on Molecular Plant-Microbe Interactions: Quebec City, Quebec, Canada, July 19-23, 2009 (Biology of plant-microbe interactions; Vol. 7). International Society for Molecular Plant-Microbe Interactions. [details]
- van Ooijen, G., Lukasik, E., van den Burg, H. A., Vossen, J. H., Cornelissen, B. J. C., & Takken, F. L. W. (2010). The small heat shock protein 20 RSI2 interacts with and is required for stability and function of tomato resistance protein I-2. Plant Journal, 63(4), 563-572. https://doi.org/10.1111/j.1365-313X.2010.04260.x [details]
- van den Burg, H. A., Kini, R. K., Schuurink, R. C., & Takken, F. L. W. (2010). Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. The Plant Cell, 22(6), 1998-2016. https://doi.org/10.1105/tpc.109.070961 [details]
2009
- Houterman, P. M., Ma, L., van Ooijen, G., de Vroomen, M. J., Cornelissen, B. J. C., Takken, F. L. W., & Rep, M. (2009). The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant Journal, 58(6), 970-978. https://doi.org/10.1111/j.1365-313X.2009.03838.x [details]
- Lukasik, E., & Takken, F. L. W. (2009). STANDing strong, resistance proteins instigators of plant defence. Current Opinion in Plant Biology, 12(4), 427-436. https://doi.org/10.1016/j.pbi.2009.03.001 [details]
- Slootweg, E., Tameling, W., Roosien, J., Lukasik, E., Joosten, M., Takken, F., Bakker, J., & Goverse, A. (2009). An outlook on the localisation and structure-function relationships of R proteins in Solanum. Potato Research, 52(3), 229-235. https://doi.org/10.1007/s11540-009-9130-9 [details]
- Takken, F. L. W., & Tameling, W. I. L. (2009). To nibble at plant resistance proteins. Science, 324(5928), 744-746. https://doi.org/10.1126/science.1171666 [details]
- van den Burg, H. A., & Takken, F. L. W. (2009). Does chromatin remodeling mark systemic acquired resistance? Trends in Plant Science, 14(5), 286-294. https://doi.org/10.1016/j.tplants.2009.02.003 [details]
2008
 Tameling, W. I. L., & Takken, F. L. W. (2008). Resistance proteins: Scouts of the plant innate immune system. European Journal of Plant Pathology, 121(3), 243-255. https://doi.org/10.1007/s10658-007-9187-8 [details]
Tameling, W. I. L., & Takken, F. L. W. (2008). Resistance proteins: Scouts of the plant innate immune system. European Journal of Plant Pathology, 121(3), 243-255. https://doi.org/10.1007/s10658-007-9187-8 [details]- van Ooijen, G., Mayr, G., Albrecht, M., Cornelissen, B. J. C., & Takken, F. L. W. (2008). Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Molecular Plant, 1(3), 401-410. https://doi.org/10.1093/mp/ssn009 [details]
- van Ooijen, G., Mayr, G., Kasiem, M. M. A., Albrecht, M., Cornelissen, B. J. C., & Takken, F. L. W. (2008). Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. Journal of Experimental Botany, 59(6), 1383-1397. https://doi.org/10.1093/jxb/ern045 [details]
- van den Burg, H. A., Tsitsigiannis, D. I., Rowland, O., Lo, J., Rallapalli, G., MacLean, D., Takken, F. L. W., & Jones, J. D. G. (2008). The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. The Plant Cell, 20(3), 697-719. https://doi.org/10.1105/tpc.107.056978 [details]
2007
- Gabriels, S. H. E. J., Vossen, J. H., Ekengren, S. K., van Ooijen, G., Abd-El-Haliem, A. M., van den Berg, G. C. M., Rainey, D. Y., Martin, G. B., Takken, F. L. W., de Wit, P. J. G. M., & Joosten, M. H. A. J. (2007). An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant Journal, 50(1), 14-28. https://doi.org/10.1111/j.1365-313X.2007.03027.x [details]
- van Ooijen, G., van den Burg, H. A., Cornelissen, B. J. C., & Takken, F. L. W. (2007). Structure and function of Resistance proteins in solanaceous plants. Annual Review of Phytopathology, 45, 43-72. https://doi.org/10.1146/annurev.phyto.45.062806.094430 [details]
2006
- Albrecht, M., & Takken, F. L. W. (2006). Update on the domain architectures of NLRs and R proteins. Biochemical and Biophysical Research Communications, 339(2), 459-462. Advance online publication. https://doi.org/10.1016/j.bbrc.2005.10.074 [details]
- Gabriels, S. H. E. J., Takken, F. L. W., Vossen, J. H., de Jong, C. F., Liu, Q., Turk, S. C. H. J., Wachowski, L. K., Peters, J., Witsenboer, H. M. A., de Wit, P. J. G. M., & Joosten, M. H. A. J. (2006). cDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Molecular Plant-Microbe Interactions, 19, 567-576. https://doi.org/10.1094/MPMI-19-0567 [details]
- Takken, F. L. W., Albrecht, M., & Tameling, W. I. L. (2006). Resistance proteins: molecular switches of plant defence. Current Opinion in Plant Biology, 9, 383-390. https://doi.org/10.1016/j.pbi.2006.05.009 [details]
- Takken, F. L. W., Tameling, W. I. L., Vossen, J. H., Albrecht, M., Berden, J. A., van Ooijen, G., & Cornelissen, B. J. C. (2006). The NB-ARC domain of R proteins acts as a molecular switch regulating plant innate immunity. In Q. C. Lópes-Lara Sánchez, S, & Geiger, O. (Eds.), Biology of Plant-Microbe Interactions. (pp. 225-230) [details]
- Tameling, W. I. L., Vossen, J. H., Albrecht, M., Lengauer, T., Berden, J. A., Haring, M. A., Cornelissen, B. J. C., & Takken, F. L. W. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiology, 140, 1233-1245. https://doi.org/10.1104/pp.105.073510 [details]
2005
- de la Fuente van Bentem, S., Vossen, J. H., de Vries, K. J., van Wees, A. C. M., Tameling, W. I. L., Dekker, H. L., de Koster, C. G., Haring, M. A., Takken, F. L. W., & Cornelissen, B. J. C. (2005). Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant Journal, 43, 284-298. https://doi.org/10.1111/j.1365-313X.2005.02450.x [details]
2004
- Takken, F. L. W., van Wijk, R., Michielse, C. B., Houterman, P. M., Ram, A. F., & Cornelissen, B. J. C. (2004). A one-step method to convert vectors into binary vectors suited for Agrobacterium-mediated transformation. Current Genetics, 45, 242-248. https://doi.org/10.1007/s00294-003-0481-5 [details]
2002
- Luderer, R., Takken, F. L. W., de Wit, P. J. G. M., & Joosten, M. H. A. J. (2002). Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Molecular Microbiology, 45(3), 875-884. https://doi.org/10.1046/j.1365-2958.2002.03060.x [details]
- Tameling, W. I. L., Elzinga, S. D. J., Darmin, P. S., Vossen, J. H., Takken, F. L. W., Haring, M. A., & Cornelissen, B. J. C. (2002). The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. The Plant Cell, 14, 2929-2939. https://doi.org/10.1105/tpc.005793 [details]
- de Jong, C. F., Takken, F. L. W., Cai, X., de Wit, P. J. G. M., & Joosten, M. H. A. J. (2002). Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Molecular Plant-Microbe Interactions, 15(10), 1040-1049. https://doi.org/10.1094/MPMI.2002.15.10.1040 [details]
- de Wit, P. J. G. M., Brandwagt, B. F., van den Burg, H. A., Cai, X., van der Hoorn, R. A. L., de Jong, C. F., van 't Klooster, J., de Kock, M. J. D., Kruijt, M., Lindhout, W. H., Luderer, R., Takken, F. L. W., Westerink, N., Vervoort, J. J. M., & Joosten, M. H. A. J. (2002). The molecular basis of co-evolution between Cladosporium fulvum and tomato. Antonie van Leeuwenhoek, 81, 409-412. https://doi.org/10.1023/A:1020553120889 [details]
2019
 Franken, P., Takken, F. L. W., & Rep, M. (2019). Transcript accumulation in a trifold interaction gives insight into mechanisms of biocontrol. New Phytologist, 224(2), 547-549. https://doi.org/10.1111/nph.16141 [details]
Franken, P., Takken, F. L. W., & Rep, M. (2019). Transcript accumulation in a trifold interaction gives insight into mechanisms of biocontrol. New Phytologist, 224(2), 547-549. https://doi.org/10.1111/nph.16141 [details]
2010
 van den Burg, H. A., & Takken, F. L. W. (2010). SUMO-, MAPK-, and resistance protein-signaling converge at transcription complexes that regulate plant innate immunity. Plant Signaling & Behavior, 5(12), 1587-1591. https://doi.org/10.4161/psb.5.12.13913 [details]
van den Burg, H. A., & Takken, F. L. W. (2010). SUMO-, MAPK-, and resistance protein-signaling converge at transcription complexes that regulate plant innate immunity. Plant Signaling & Behavior, 5(12), 1587-1591. https://doi.org/10.4161/psb.5.12.13913 [details]
2018
- Takken, F. L. W., van den Burg, H. A., & Di, X. (2018). Plants comprising pathogen effector constructs. (Patent No. pct/nl21017/050798).
2008
- Tameling, W. I. L., & Takken, F. L. W. (2008). Resistance proteins: Scouts of the plant innate immune system. In D. B. Collinge, L. Munk, & B. M. Cooke (Eds.), Sustainable disease management in a European context (pp. 243-255). Springer. https://doi.org/10.1007/978-1-4020-8780-6_4 [details]
2005
- Takken, F. L. W. (2005). Overeenkomsten en verschillen in ziekteresistentie tussen plant en dier. Gewasbescherming, 36, 262. [details]
2004
- Joosten, M. H. A. J., de Jong, C. F., de Wit, P. J. G. M., Takken, F. L. W., & Turk, S. C. H. J. (2004). Nucleotide sequences involved in plant resistance.
2003
- Turk, S. C. H. J., Takken, F. L. W., de Jong, C. F., Joosten, M. H. A. J., & de Wit, P. J. G. M. (2003). Nucleotide sequences involved in plant disease resistance.
2002
- Takken, F. L. W., & de Wit, P. J. G. M. (2002). Elicitor from Cladosporium.
Talk / presentation
- Takken, F. L. W. (speaker) (19-5-2011). The role of pathogen effectors in NLR-mediated plant innate immunity, International Meeting of the Collaborative Research Center SFB 648 “ Communication in Plants and their Responses to the Environment”, Halle (Saale), Germany.
- Takken, F. L. W. (speaker) (8-10-2010). Molecular co-evolution in the tomato-Fusarium pathosystem, Justus Liebig University , Giessen, Germany, Giessen, Germany.
- Takken, F. L. W. (invited speaker) (8-6-2010). Functional analysis of Fusarium oxysporum f.sp. lycopersici effector proteins., DFG funded colloquium: 'Microbial reprogramming of plant cell development' priority project SPP1212 'Plant-Micro', Freising, Germany.
- Takken, F. L. W. (speaker) (11-5-2010). The arms race between effectors of Fusarium oxysporum and resistance proteins of tomato., Seminar series School of Biological and Biomedical Sciences, Durham University,, Durham, UK.
- Takken, F. L. W. (speaker) (1-8-2009). How to resist a resistance protein?, British Society for Plant Pathology, Presidential Meeting 2009, Oxford, UK.
- Takken, F. L. W. (speaker) (19-7-2009). How to resist tomato resistance proteins?, XIV International Congress on Molecular Plant-MicrobeInteractions, Quebec, Canada.
- Takken, F. L. W. (speaker) (22-9-2008). Structure-function analysis of plant NB-LRR disease resistance proteins at the SFB 670, Structure Function and Evolution of Innate immunity, Max-Planck-Institut für Züchtungsforschung Koln, Germany.
- Takken, F. L. W. (speaker) (9-9-2008). Resistance proteins: scouts of the plant innate immune system, 18th EUCARPIA congress, "Modern Variety Breeding for Present and Future Needs, Valencia, Spain.
- Takken, F. L. W. (speaker) (25-6-2008). Resistance proteins: scouts of the plant innate immune system., 4th EPSO Conference, Presqu'ile de Giens (Côte d'Azur), France.
- Takken, F. L. W. (speaker) (18-6-2008). Resistance proteins: scouts of the plant innate immune system., EPS/Bioexploit Summerschool “On the evolution of plant-pathogen interactions: from principles to practices, Wageningen, the Netherlands.
- Takken, F. L. W. (speaker) (1-5-2008). Molecular aspects of I-2 mediated resistance to Fusarium oxysporum., Max-Planck-Institut für Züchtungsforschung, Koln, Germany.
- Takken, F. L. W. (speaker) (10-2-2008). Function of the Nucleotide Binding Domain for R Proteins, Keystone Symposium on Plant Innate Immunity, Resort in Keystone, Colorado, USA.
- Takken, F. L. W. (speaker) (7-12-2007). Molecular mechanisms regulating the activity of plant NB-LRR resistance proteins., seminar: Institute of Plant Biology., Zurich, Switzerland.
- Takken, F. L. W. (speaker) (20-11-2007). The NB-ARC domain of R proteins acts as a molecular switch regulating plant innate immunity., seminar: Max von Pettenkofer Institut., LMU München, Germany.
- Lukasik, E. (speaker), Cornelissen, B. J. C. (speaker) & Takken, F. L. W. (speaker) (17-10-2007). Role of nucleotide binding for R protein function., Bioexploit Meeting., Lunteren, the Netherlands.
- Takken, F. L. W. (speaker) (12-7-2007). Host-Pathogen Interaction from Plants to Mammals -Distinct and Shared Pathways of Immune Defence., seminar: Inflammatory diseases of barrier organs., University Hospital, Kiel, Germany.
- van Ooijen, G. (speaker), Cornelissen, B. J. C. (speaker) & Takken, F. L. W. (speaker) (1-1-2007). A role for the conserved MHD motif for R protein function., XIIIth International Congress on Molecular Plant-Microbe Interactions., Sorrento, Italy.
- Takken, F. L. W. (speaker) (27-11-2006). The NB-ARC domain: an NTP-hydrolysing molecular switch., Comparative Biology of Innate Immune Systems, New York, USA.
- Takken, F. L. W. (speaker), Tameling, W. I. L. (speaker), Albrecht, M. (speaker), van Ooijen, G. (speaker), de Vroomen, M. J. (speaker), de Vries, K. J. (speaker) & Cornelissen, B. J. C. (speaker) (13-8-2006). R proteins, molecular switches of plant defence., 8th Conference of the European Foundation for Plant Pathology & British Society of Plant Pathology Presedential Meeting, Copenhagen, Denmark.
- Takken, F. L. W. (speaker) (2-2-2006). Innate immune receptors in plants and animals; do anlogous modules have comparable functions?, EPS Symposium Intracellular signalling, UvA, Amsterdam, The Netherlands.
- Takken, F. L. W. (speaker), Tameling, W. I. L. (speaker), Vossen, J. H. (speaker), Albrecht, M. (speaker), Berden, J. A. (speaker), Haring, M. A. (speaker) & Cornelissen, B. J. C. (speaker) (14-12-2005). The NB-ARC domain of R proteins acts as a molecular switch regulating plant innate immunity., XII Int. congress on Molecular-Plant Microbe Interactions., Merida, Mexico.
- Takken, F. L. W. (speaker) (27-4-2005). Overeenkomsten en verschillen in ziekteresistentie tussen plant en dier., Meeting: In gewasbeschermingsmanifestatie: Is het al tijd om te oogsten., Ede, The Netherlands.
- Takken, F. L. W. (speaker) (27-4-2005). R proteins: molecular switches of disease resistance signalling., In 4e Gewasbeschermingsmanifestatie, Ede , The Netherlands.
- Takken, F. L. W. (speaker) (4-4-2005). The role of nucleotide binding for R protein function., EPS Symposium, Lunteren, The Netherlands.
- Takken, F. L. W. (speaker) (26-7-2004). R proteins: Molecular switches of disease resistance signalling., Invitation from the Boyce Thompson Institute at the Cornell University, Ithaca, United States.
- Takken, F. L. W. (speaker) (6-4-2004). R proteins: molecular switches of disease resistance signalling., ALW/EPW Meeting, Lunteren, Nederland.
- Takken, F. L. W. (speaker) (11-3-2004). R proteins: molecular switches of disease resistance signalling., 10th Nederland Biotechnology Conference, Ede, Nederland.
- Takken, F. L. W. (speaker) (10-4-2003). Elucidation of the molecular and biochemical basis of I-2 mediated Fusarium resistance, Keystone Plant Biology: Functions and control of cell death - symposium, Snowbird, Utah, VS.
- Takken, F. L. W. (speaker) (30-1-2003). The molecular and biochemical basis of I-2 mediated Fusarium resistance in tomato, WCS dat, Utrecht.
2023
2020
2017
2014
2012
2011
2017
- Di, X., Cao, L., Hughes, R. K., Tintor, N., Banfield, M. J. & Takken, F. L. W. (16-8-2017). Avr2 effector protein from the fungal plant pathogen Fusarium oxysporum. Protein Data Bank (PDB). https://doi.org/10.2210/pdb5od4/pdb
This list of publications is extracted from the UvA-Current Research Information System. Questions? Ask the library or the Pure staff of your faculty / institute. Log in to Pure to edit your publications. Log in to Personal Page Publication Selection tool to manage the visibility of your publications on this list. -
Ancillary activities
- No ancillary activities