Research lines Bacterial Cell Biology & Physiology Groups
Bacterial Cell Biology
Research line Prof. Leendert Hamoen
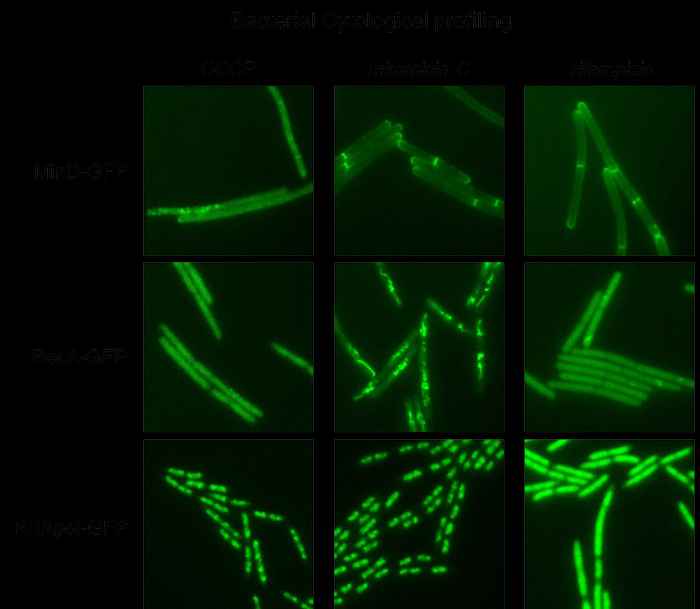
Cell division is a vital process, and yet, we still do not know exactly how bacterial cells divide. We use the well-known bacterial model system Bacillus subtilis to study the dynamic interaction between cell division proteins using molecular biological and fluorescence microscopy and techniques. This latter technique is also very useful to study the mode of action of novel antimicrobial compounds, which is called bacterial cytological profiling. We this and other tools to identify and characterize new antimicrobial compounds.
B. subtilis is used in the bioindustry to produce enzymes and vitamins. The bacterium is also used as a probiotic in animal feed, and in fact it is eaten by humans in the form of Natto. In collaboration with industrial partners we help to further optimize B. subtilis for applied purposes.
Bacterial Cell Biology
Research line Dr. Gaurav Dugar
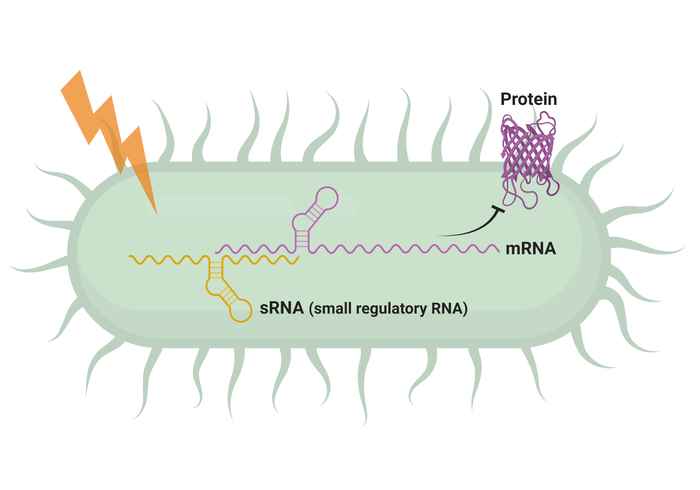
Regulatory small RNAs (sRNAs) play a crucial role in regulating gene expression in several cellular processes in bacteria, contributing to their adaptability and survival. Meanwhile, the organization of the bacterial chromosome also impacts gene regulation, as the spatial arrangement of genes influences their accessibility and interactions. By harnessing the power of new genome-wide capture techniques to capture interaction between nucleic acids genome-wide combined with unsupervised learning methods, we aim to uncover hidden patterns and relationships within these fundamental regulatory processes. Our findings can provide valuable insights into how bacteria adapt and respond to different environments. By exploring the intricate connections within vast datasets, we aim to contribute to a deeper understanding of bacterial gene regulation.
Molecular Microbial Physiology
Research line Dr Filipe Branco dos Santos
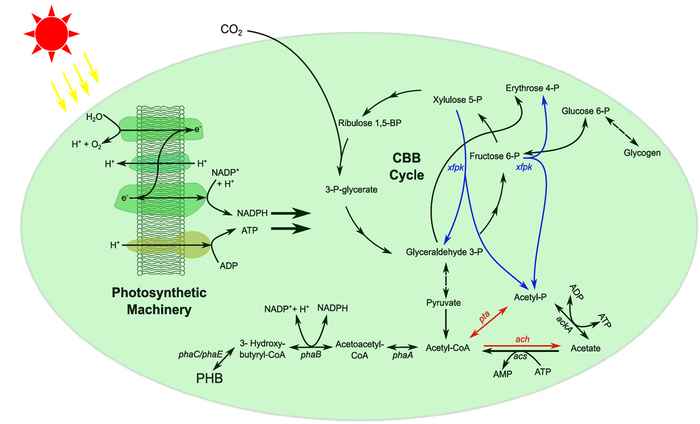
We aim to integrate knowledge on the biochemical and biophysical properties of signal transduction- and metabolic networks that regulate and underlie fermentation, respiration and oxygenic photosynthesis. Ultimately, we want to understand how the environment, through evolution and natural selection, shapes microbes to be equipped with the capacity to deal with the fluctuations they encounter. For our studies, we focus on the industrially relevant lactic acid bacterium Lactococcus lactis and the model organism for studies in oxygenic photosynthesis Synechocystis PCC6803. Both organisms are highly relevant for the transition of our society towards a sustainable bio-based economy.